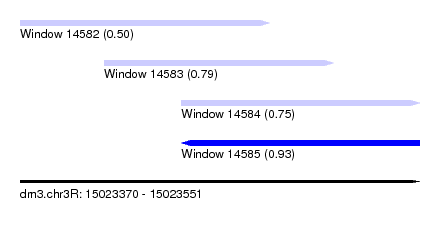
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,023,370 – 15,023,551 |
| Length | 181 |
| Max. P | 0.934690 |

| Location | 15,023,370 – 15,023,483 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
| Shannon entropy | 0.47560 |
| G+C content | 0.54241 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.94 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
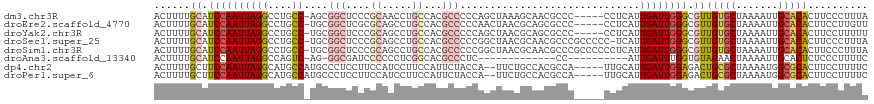
>dm3.chr3R 15023370 113 + 27905053 ACUUUUGCAUCCAAUUAGGCCUGCC-AGCGGCUCCCGCAACCUGCCACGCCCCCAGCUAAAGCAACGCCC-----CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUA .....((((........(((.((.(-((..((....))...))).)).)))...(((....(((((((((-----..((...))..)))))))))))).....))))............ ( -30.90, z-score = -1.47, R) >droEre2.scaffold_4770 11136688 113 - 17746568 ACUUUUGCAUCCAAUUAGGCCUGCC-UGCGGCUCGCGCAGCCUGCCACGCCCCCAACUAACGCAGCGCCC-----CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUGUU .....((((........(((..(.(-((((.....))))).).)))..............((((((((((-----..((...))..)))))))))).......))))............ ( -33.50, z-score = -0.99, R) >droYak2.chr3R 3712050 113 + 28832112 ACUUUUGCAUCCAAUUAGGCCUGCC-UGCGGCUCCCGCAGCCUGCCACGCCCCCAGCUAACGCAGCGCCC-----CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUUUU .....((((........(((..(.(-(((((...)))))).).)))..............((((((((((-----..((...))..)))))))))).......))))............ ( -35.30, z-score = -2.01, R) >droSec1.super_25 85745 117 - 827797 ACUUUUGCAUCCAAUUAGGCCUGCC-UGCGGCUCCCGCAGCCUGCCACGCCCCCGGCUAACGCAACGCCCGCCCCC-UCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUA .....((((........(((..(.(-(((((...)))))).).)))..(((...)))...(((((((((((.....-........))))))))))).......))))............ ( -37.32, z-score = -2.28, R) >droSim1.chr3R 21058808 118 - 27517382 ACUUUUGCAUCCAAUUAGGCCUGCC-UGCGGCUCCCGCAGCCUGCCACGCCCCCGGCUAACGCAACGCCCGCCCCCCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUA .....((((........(((..(.(-(((((...)))))).).)))..(((...)))...(((((((((((..(........)..))))))))))).......))))............ ( -37.60, z-score = -2.34, R) >droAna3.scaffold_13340 8334814 94 - 23697760 ACUUUUGCAUCCAAUUAGGCCAGUC-AG-GGCGAUCCCCCCUCGGCACGCCCUC-------------CC----------AUUGAUUUGGUGUAGAACUAAAAUUGCACUCUCCCUUUUC ..(((((((((.((((((....(..-((-((((..((......))..)))))).-------------.)----------.)))))).)))))))))....................... ( -22.80, z-score = -1.22, R) >dp4.chr2 29468824 112 + 30794189 ACUUUUGCUUCCAAUUAGGCAUGCCAUGCCCUCCUUCCAUCCUUCCAUUCUACCA--UUCUGCCACGCCA-----UUGCAUUGAUUGGAGACUGCGCUAAAAUGGCGCACUUCCUUUUC ........((((((((((((((...))))).........................--.........((..-----..))..)))))))))..((((((.....)))))).......... ( -22.70, z-score = -1.31, R) >droPer1.super_6 4839460 112 + 6141320 ACUUUUGCUUCCAAUUAGGCAUGCCAUGCCCUCCUUCCAUCCUUCCAUUCUACCA--UUCUGCCACGCCA-----UUGCAUUGAUUGGAGACUGCGCUAAAAUGGCGCACUUCCUUUUC ........((((((((((((((...))))).........................--.........((..-----..))..)))))))))..((((((.....)))))).......... ( -22.70, z-score = -1.31, R) >consensus ACUUUUGCAUCCAAUUAGGCCUGCC_UGCGGCUCCCGCAGCCUGCCACGCCCCCAGCUAACGCAACGCCC_____CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUUUA ......((.((((((((((((........))))...((.....))....................................)))))))).))(((((.......))))).......... (-13.59 = -13.94 + 0.34)
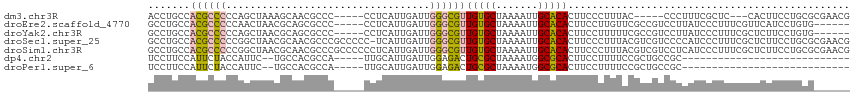
| Location | 15,023,408 – 15,023,512 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.11 |
| Shannon entropy | 0.58862 |
| G+C content | 0.56538 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -10.35 |
| Energy contribution | -9.78 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15023408 104 + 27905053 ACCUGCCACGCCCCCAGCUAAAGCAACGCCC-----CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUAC-----CCCUUUCGCUC---CACUUCCUGCGCGAACG ........(((...(((....((((((((((-----..((...))..)))))))((((.......))))............-----.......))).---......))).))).... ( -23.40, z-score = -2.65, R) >droEre2.scaffold_4770 11136726 106 - 17746568 GCCUGCCACGCCCCCAACUAACGCAGCGCCC-----CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUGUUCGCCGUCCUUAUCCCUUUCGUUCAUCCUGUG------ ......((((.....(((...((((((((((-----..((...))..))))))))))........((..((......))..))................))).....))))------ ( -20.00, z-score = -0.99, R) >droYak2.chr3R 3712088 106 + 28832112 GCCUGCCACGCCCCCAGCUAACGCAGCGCCC-----CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUUUUCGCCGUCCUUAUCCCUUUCGCUCUUCCUGUG------ ......((((.....(((....(((((((((-----..((...))..)))))))))((.......))................................))).....))))------ ( -21.20, z-score = -1.18, R) >droSec1.super_25 85783 116 - 827797 GCCUGCCACGCCCCCGGCUAACGCAACGCCCGCCCCC-UCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUACGUCGUCCCCAUCCCUUUCGCUCUUCCUGCGCGAACG ((..(((........)))...(((((((((((.....-........)))))))))))........)).............................(((((.(.....).))))).. ( -28.02, z-score = -2.00, R) >droSim1.chr3R 21058846 117 - 27517382 GCCUGCCACGCCCCCGGCUAACGCAACGCCCGCCCCCCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUACGUCGUCCUCAUCCCUUUCGCUCUUCCUGCGCGAACG ((..(((........)))...(((((((((((..(........)..)))))))))))........)).............................(((((.(.....).))))).. ( -28.30, z-score = -2.15, R) >dp4.chr2 29468863 82 + 30794189 UCCUUCCAUUCUACCAUUC--UGCCACGCCA-----UUGCAUUGAUUGGAGACUGCGCUAAAAUGGCGCACUUCCUUUUCCGCUGCCGC---------------------------- ...................--..........-----..(((..((..((((..((((((.....)))))).))))...))...)))...---------------------------- ( -15.00, z-score = 0.05, R) >droPer1.super_6 4839499 82 + 6141320 UCCUUCCAUUCUACCAUUC--UGCCACGCCA-----UUGCAUUGAUUGGAGACUGCGCUAAAAUGGCGCACUUCCUUUUCCGCUGCCGC---------------------------- ...................--..........-----..(((..((..((((..((((((.....)))))).))))...))...)))...---------------------------- ( -15.00, z-score = 0.05, R) >consensus GCCUGCCACGCCCCCAGCUAACGCAACGCCC_____CCUCAUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUUUACGCCGUCCCUAUCCCUUUCGCUCUUCCUGCG______ .......((((((..................................))))))(((((.......)))))............................................... (-10.35 = -9.78 + -0.57)
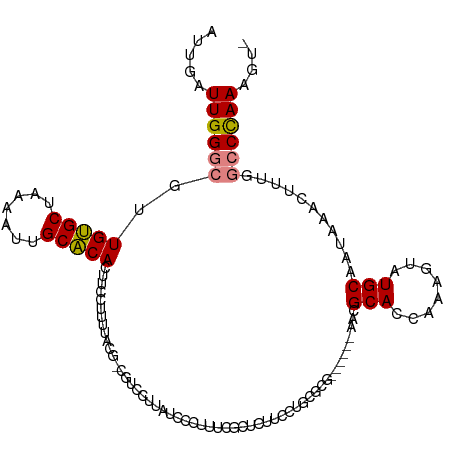
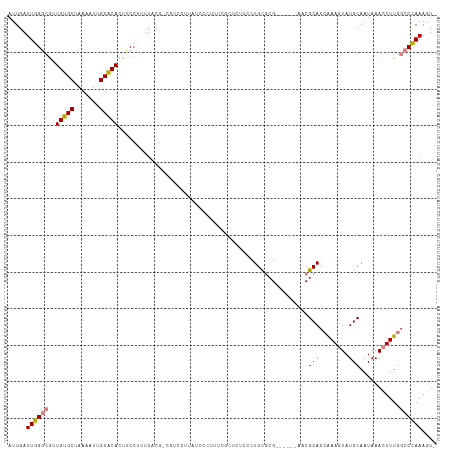
| Location | 15,023,443 – 15,023,551 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.53 |
| Shannon entropy | 0.45617 |
| G+C content | 0.47269 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.09 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
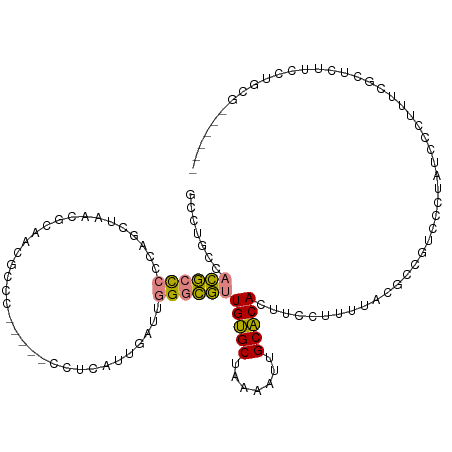
>dm3.chr3R 15023443 108 + 27905053 AUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUAC-----CCCUUUCGCUC---CACUUCCUGCGCGAACGCGAACGCACCGAAGUAUGCAAUAAACUUUGGCCCAAAGU- .....(((((((.....)....((((((.(((((.......-----....(((((.(---........).))))).((....))...))))).))))))........))))))...- ( -27.60, z-score = -1.78, R) >droEre2.scaffold_4770 11136761 110 - 17746568 AUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUGUUCGCCGUCCUUAUCCCUUUCGUUCAUCCUGUGCG------AACGCACCAAACUAUGCAAUAAACUUUGGCCCAAAGU- .....((((((..(((((.......))))).......((((((((......................)).)))------)))(((........)))...........))))))...- ( -25.25, z-score = -1.55, R) >droYak2.chr3R 3712123 110 + 28832112 AUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUUUUCGCCGUCCUUAUCCCUUUCGCUCUUCCUGUGCG------AACGCACCAAACUAUGCAAUAAACUUUGGCCCAAAGU- .....((((((..(((((.......)))))..........................(((((.(.....).)))------)).(((........)))...........))))))...- ( -24.40, z-score = -1.68, R) >droSec1.super_25 85822 116 - 827797 AUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUACGUCGUCCCCAUCCCUUUCGCUCUUCCUGCGCGAACGCGAACGCAGCGAAGUAUGCAAUAAACUUUGGCCCAAAGU- .....((((((..(((((.......))))).............................((.(((((((((((....))..))))).))))...))...........))))))...- ( -31.10, z-score = -1.49, R) >droSim1.chr3R 21058886 116 - 27517382 AUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCCUUUACGUCGUCCUCAUCCCUUUCGCUCUUCCUGCGCGAACGCGAACGCACCGAAGUAUGCAAUAAACUUUGGCCCAAAGU- .....(((((((((((((.......))))...........((.(((...........((((.(.....).)))))))))))))).(((((((.........))))))).))))...- ( -29.00, z-score = -1.32, R) >dp4.chr2 29468896 85 + 30794189 AUUGAUUGGAGACUGCGCUAAAAUGGCGCACUUCCUUUUCCG------------------------CUGC-CG-------CCGCACAAAAGUAUGCAAUAAACUUUCGGCUAAACUU ...((..((((..((((((.....)))))).))))...))..------------------------....-.(-------(((....(((((.........)))))))))....... ( -19.20, z-score = -0.41, R) >droPer1.super_6 4839532 85 + 6141320 AUUGAUUGGAGACUGCGCUAAAAUGGCGCACUUCCUUUUCCG------------------------CUGC-CG-------CCGCACAAAAGUAUGCAAUAAACUUUCGGCUAAACUU ...((..((((..((((((.....)))))).))))...))..------------------------....-.(-------(((....(((((.........)))))))))....... ( -19.20, z-score = -0.41, R) >consensus AUUGAUUGGGCGUUGUGCUAAAAUUGCACACUUCCUUUUACG_CGUCCUUAUCCCUUUCGCUCUUCCUGCGCG______AACGCACCAAAGUAUGCAAUAAACUUUGGCCCAAAGU_ .....((((((..(((((.......)))))....................................................(((........)))...........)))))).... (-16.12 = -16.09 + -0.04)
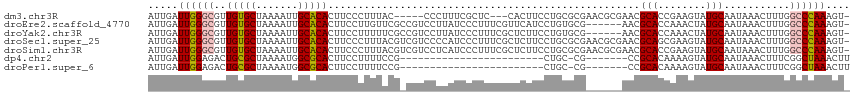
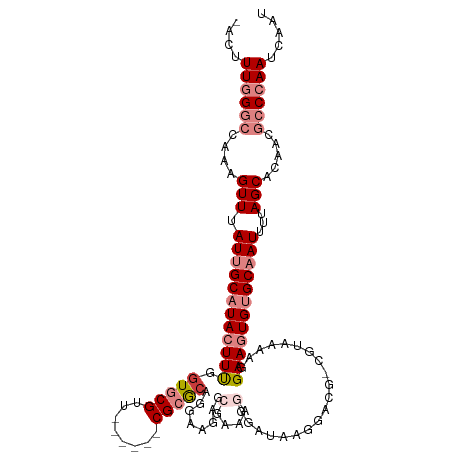
| Location | 15,023,443 – 15,023,551 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.53 |
| Shannon entropy | 0.45617 |
| G+C content | 0.47269 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -20.65 |
| Energy contribution | -22.56 |
| Covariance contribution | 1.91 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
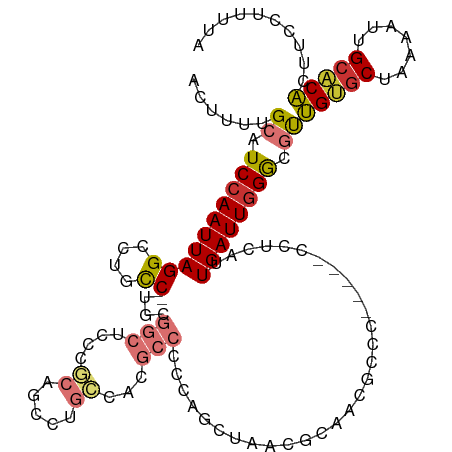
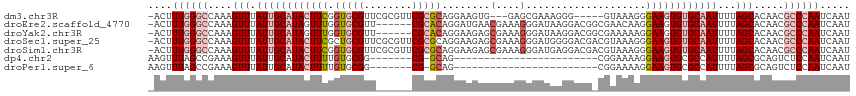
>dm3.chr3R 15023443 108 - 27905053 -ACUUUGGGCCAAAGUUUAUUGCAUACUUCGGUGCGUUCGCGUUCGCGCAGGAAGUG---GAGCGAAAGGG-----GUAAAGGGAAGUGUGCAAUUUUAGCACAACGCCCAAUCAAU -...((((((....(((.((((((((((((..(((.(((((.(((((.......)))---)))))))....-----)))....))))))))))))...))).....))))))..... ( -38.80, z-score = -2.86, R) >droEre2.scaffold_4770 11136761 110 + 17746568 -ACUUUGGGCCAAAGUUUAUUGCAUAGUUUGGUGCGUU------CGCACAGGAUGAACGAAAGGGAUAAGGACGGCGAACAAGGAAGUGUGCAAUUUUAGCACAACGCCCAAUCAAU -...((((((....(((.((((((((.(((.....(((------(((..........(....)...........))))))...))).))))))))...))).....))))))..... ( -30.70, z-score = -1.57, R) >droYak2.chr3R 3712123 110 - 28832112 -ACUUUGGGCCAAAGUUUAUUGCAUAGUUUGGUGCGUU------CGCACAGGAAGAGCGAAAGGGAUAAGGACGGCGAAAAAGGAAGUGUGCAAUUUUAGCACAACGCCCAAUCAAU -...((((((.........((((...((((.((((...------.))))........(....)......)))).)))).........(((((.......)))))..))))))..... ( -28.70, z-score = -0.95, R) >droSec1.super_25 85822 116 + 827797 -ACUUUGGGCCAAAGUUUAUUGCAUACUUCGCUGCGUUCGCGUUCGCGCAGGAAGAGCGAAAGGGAUGGGGACGACGUAAAGGGAAGUGUGCAAUUUUAGCACAACGCCCAAUCAAU -...((((((....(((.((((((((((((.((.(((((.(((((.(((.......)))....))))).)))))......)).))))))))))))...))).....))))))..... ( -44.90, z-score = -3.73, R) >droSim1.chr3R 21058886 116 + 27517382 -ACUUUGGGCCAAAGUUUAUUGCAUACUUCGGUGCGUUCGCGUUCGCGCAGGAAGAGCGAAAGGGAUGAGGACGACGUAAAGGGAAGUGUGCAAUUUUAGCACAACGCCCAAUCAAU -...((((((....(((.((((((((((((....(((((.(((((.(((.......)))....))))).))))).(.....).))))))))))))...))).....))))))..... ( -39.50, z-score = -2.45, R) >dp4.chr2 29468896 85 - 30794189 AAGUUUAGCCGAAAGUUUAUUGCAUACUUUUGUGCGG-------CG-GCAG------------------------CGGAAAAGGAAGUGCGCCAUUUUAGCGCAGUCUCCAAUCAAU .......((((........(((((((....)))))))-------))-))..------------------------.......((((.(((((.......))))).).)))....... ( -21.70, z-score = -0.03, R) >droPer1.super_6 4839532 85 - 6141320 AAGUUUAGCCGAAAGUUUAUUGCAUACUUUUGUGCGG-------CG-GCAG------------------------CGGAAAAGGAAGUGCGCCAUUUUAGCGCAGUCUCCAAUCAAU .......((((........(((((((....)))))))-------))-))..------------------------.......((((.(((((.......))))).).)))....... ( -21.70, z-score = -0.03, R) >consensus _ACUUUGGGCCAAAGUUUAUUGCAUACUUUGGUGCGUU______CGCGCAGGAAGAGCGAAAGGGAUAAGGACG_CGUAAAAGGAAGUGUGCAAUUUUAGCACAACGCCCAAUCAAU ....((((((....(((.((((((((((((.(((((........)))))........(....)....................))))))))))))...))).....))))))..... (-20.65 = -22.56 + 1.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:08 2011