
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,012,606 – 15,012,696 |
| Length | 90 |
| Max. P | 0.681983 |

| Location | 15,012,606 – 15,012,696 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.47966 |
| G+C content | 0.37176 |
| Mean single sequence MFE | -8.51 |
| Consensus MFE | -6.18 |
| Energy contribution | -6.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.681983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
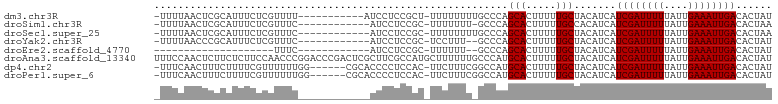
>dm3.chr3R 15012606 90 + 27905053 -UUUUAACUCGCAUUUCUCGUUUU-----------AUCCUCCGCU-UUUUUUUUGCCCAGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU -.......................-----------.......((.-........))..((((.....))))......((((((((....))))))))...... ( -7.60, z-score = -0.69, R) >droSim1.chr3R 21052427 88 - 27517382 -UUUUAACUCGCAUUUCUCGUUUC------------AUCCUCCGC-UUUUUUU-GCCCAGCACUUUUUGCCACAUCAUCGAUUUUUAUUGAAAUUGACACUAA -.......................------------.......((-.......-))...(((.....))).......((((((((....))))))))...... ( -6.50, z-score = -0.06, R) >droSec1.super_25 79454 89 - 827797 -UUUUAACUCGCAUUUCUCGUUUC------------AUCCUCCGC-UUUUUUUUGCCCAGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAA -.......................------------.......((-........))..((((.....))))......((((((((....))))))))...... ( -8.40, z-score = -0.95, R) >droYak2.chr3R 3699011 87 + 28832112 -UUUUAACCCGCAUUUCUCGUUUC------------AUCCUCCGC-UCCUUU--GCCCAGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU -.......................------------.......((-......--))..((((.....))))......((((((((....))))))))...... ( -8.60, z-score = -1.27, R) >droEre2.scaffold_4770 11130328 68 - 17746568 --------------------UUUC------------AUCCUCCGC-UUUUUU--GCCCAGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU --------------------....------------.......((-......--))..((((.....))))......((((((((....))))))))...... ( -8.60, z-score = -2.31, R) >droAna3.scaffold_13340 8328514 103 - 23697760 UUUCCAACUCUUCUCUUCCAACCCGGACCCGACUCGCUUCGCCAUGCUUUUUUGCCCAUGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU ................(((.....))).............((...((......))....))................((((((((....))))))))...... ( -9.30, z-score = -0.08, R) >dp4.chr2 29462183 95 + 30794189 -UUUCAACUUUCUUUUCGUUUUUUGG------CGCACCCCUCCAC-UUCUUUCGGCCAUGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU -......................(((------(............-........)))).(((.....))).......((((((((....))))))))...... ( -9.55, z-score = -0.36, R) >droPer1.super_6 4832744 95 + 6141320 -UUUCAACUUUCUUUUCGUUUUUUGG------CGCACCCCUCCAC-UUCUUUCGGCCAUGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU -......................(((------(............-........)))).(((.....))).......((((((((....))))))))...... ( -9.55, z-score = -0.36, R) >consensus _UUUUAACUCGCAUUUCUCGUUUC____________AUCCUCCGC_UUUUUUU_GCCCAGCACUUUUUGCUACAUCAUCGAUUUUUAUUGAAAUUGACACUAU ...........................................................(((.....))).......((((((((....))))))))...... ( -6.18 = -6.18 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:05 2011