| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,010,911 – 15,011,034 |
| Length | 123 |
| Max. P | 0.820042 |

| Location | 15,010,911 – 15,011,006 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 88.23 |
| Shannon entropy | 0.19761 |
| G+C content | 0.48755 |
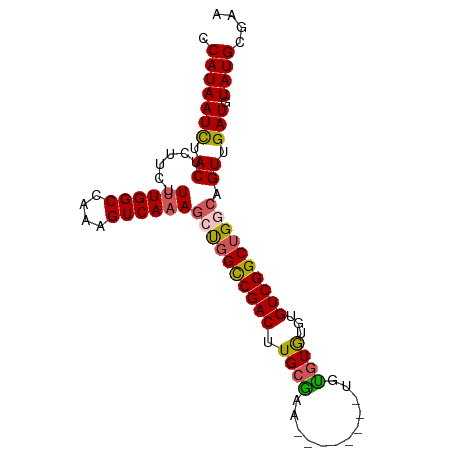
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.521818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
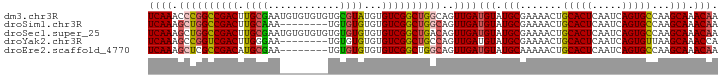
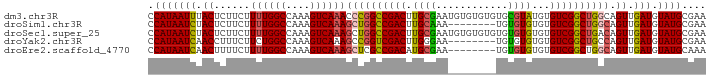
>dm3.chr3R 15010911 95 - 27905053 UCAAACCCGGCCGACUUGCGAAUGUGUGUGUGCGUAUGUGUCGGCUGGCAGUUGAUGUAUGCGAAAACUGCACUCAAUCAGUGCCAAGCAAACAA ((((..(((((((((.((((.((......)).))))...)))))))))...))))(((.(((.......(((((.....)))))...))).))). ( -33.20, z-score = -1.80, R) >droSim1.chr3R 21050658 87 + 27517382 UCAAAGCUGGCCGACUUGCAAA--------UGUGUGUGUGUCGGCUGGCAGUUGAUGUAUGCGAAAACUGCACUCAAUCAGUGCCAAGCAAACAA ((((.((..((((((.((((..--------....)))).))))))..))..))))(((.(((.......(((((.....)))))...))).))). ( -32.10, z-score = -2.55, R) >droSec1.super_25 77723 95 + 827797 UCAAAGCUGGCCGACUUGCGAAUGUGUGUGUGUGUGUGUGUCGGCUGACAGUUGAUGUAUGCGAAAACUGCACUCAAUCAGUGCCAAGCAAACAA .....((((((..(((...((.((.((((((.((((((..(((((.....)))))..))))))...)).)))).)).)))))))).)))...... ( -28.50, z-score = -0.45, R) >droYak2.chr3R 3697297 87 - 28832112 UCAAAGCCGGUCGACUUGGGAA--------UGUGUGUGUGUCGGCUGCCAGUUGAUGUAUGCGAAAACUGCACUCAAUCAGUGUUAAGCAAACCA ........(((...((((((..--------..((((((..(((((.....)))))..))))))....))(((((.....)))))))))...))). ( -19.70, z-score = 1.05, R) >droEre2.scaffold_4770 11128599 87 + 17746568 UCAAAGCUCGCCGACAUGCGAA--------UGUGUGUGUGUCGGCUGGCAGUUGAUGUAUGCAAAAACUGCACUCAAUCAGUGCCAAGCAAACAA .....(((.(((((((((((..--------....)))))))))))(((((.((((((..((((.....))))..).)))))))))))))...... ( -34.90, z-score = -3.71, R) >consensus UCAAAGCUGGCCGACUUGCGAA________UGUGUGUGUGUCGGCUGGCAGUUGAUGUAUGCGAAAACUGCACUCAAUCAGUGCCAAGCAAACAA ((((.((((((((((.((((............))))...))))))))))..))))(((.(((.......(((((.....)))))...))).))). (-23.96 = -24.12 + 0.16)
| Location | 15,010,941 – 15,011,034 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 86.64 |
| Shannon entropy | 0.22274 |
| G+C content | 0.47610 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
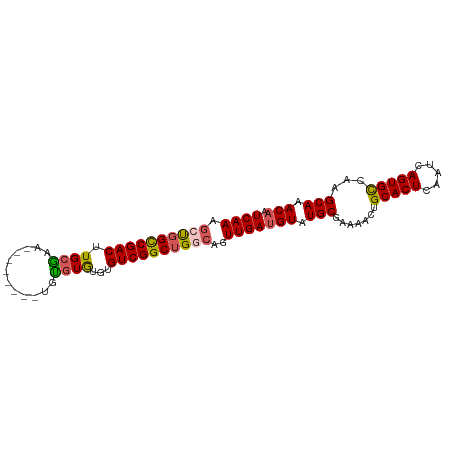
>dm3.chr3R 15010941 93 - 27905053 CCAUAAUUUACUCUUCUUUUGGCCAAAGUCAAACCCGGCCGACUUGCGAAUGUGUGUGUGCGUAUGUGUCGGCUGGCAGUUGAUGUAUGCGAA .....................((....(((((..(((((((((.((((.((......)).))))...)))))))))...)))))....))... ( -28.10, z-score = -1.51, R) >droSim1.chr3R 21050688 85 + 27517382 CCAUAAUCUACUCUUCUUUUGGCCAAAGUCAAAGCUGGCCGACUUGCAAA--------UGUGUGUGUGUCGGCUGGCAGUUGAUGUAUGCGAA .(((((((.(((.....((((((....))))))((..((((((.((((..--------....)))).))))))..))))).))).)))).... ( -29.40, z-score = -3.04, R) >droSec1.super_25 77753 93 + 827797 CCAUAAUCUACUCUUCUUUUGGCCAAAGUCAAAGCUGGCCGACUUGCGAAUGUGUGUGUGUGUGUGUGUCGGCUGACAGUUGAUGUAUGCGAA .(((((((.(((.....((((((....))))))(..(((((((.((((.(..(....)..).)))).)))))))..)))).))).)))).... ( -23.50, z-score = -0.43, R) >droYak2.chr3R 3697327 85 - 28832112 CCAUAAUCAACCUUUCUUCUGGCCAAAGUCAAAGCCGGUCGACUUGGGAA--------UGUGUGUGUGUCGGCUGCCAGUUGAUGUAUGCGAA ......((..(((.((..(((((..........)))))..))...)))..--------...(((((..(((((.....)))))..))))))). ( -22.00, z-score = -0.27, R) >droEre2.scaffold_4770 11128629 85 + 17746568 CCAUAAUCAACUUUUCUUUUGGCCAAAGUCAAAGCUCGCCGACAUGCGAA--------UGUGUGUGUGUCGGCUGGCAGUUGAUGUAUGCAAA .(((((((((((.....((((((....))))))(((.(((((((((((..--------....))))))))))).)))))))))).)))).... ( -34.20, z-score = -4.61, R) >consensus CCAUAAUCUACUCUUCUUUUGGCCAAAGUCAAAGCUGGCCGACUUGCGAA________UGUGUGUGUGUCGGCUGGCAGUUGAUGUAUGCGAA .(((((((.((......((((((....))))))((((((((((.((((............))))...)))))))))).)).))).)))).... (-21.00 = -21.16 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:04 2011