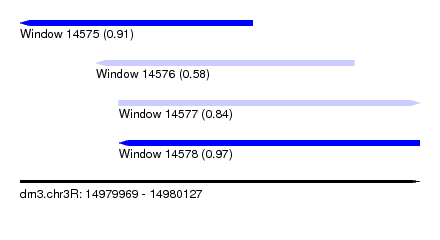
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,979,969 – 14,980,127 |
| Length | 158 |
| Max. P | 0.970013 |

| Location | 14,979,969 – 14,980,061 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.45876 |
| G+C content | 0.39875 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -10.51 |
| Energy contribution | -11.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
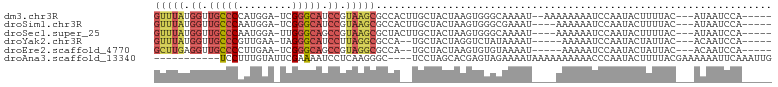
>dm3.chr3R 14979969 92 - 27905053 GUUUAUGGUUGCCCCAUGGA-UCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCAAAAU--AAAAAAAAUCCAAUACUUUUAC---AUAAUCCA----- ((((((((.(((((......-..))))).))))))))((((((((....))))).))).....--......................---........----- ( -27.30, z-score = -2.95, R) >droSim1.chr3R 21019700 90 + 27517382 GUUUAUGGUUGCCCAAUGGA-UCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCGAAAU----AAAAAAUCCAAUACUUUUAC---AUAAUCCA----- ((((((((.(((((......-..))))).))))))))((((((((....))))).))).....----....................---........----- ( -27.90, z-score = -3.11, R) >droSec1.super_25 47156 90 + 827797 GUUUAUGGUUGCCCAAUGGA-UUGGGCAGCCGUAAGCGCUACUUGCUACUAAGUGGGCAAAAU----AAAAAAUCCAAUACUUUUAC---AUAAUCCA----- ((((((((((((((((....-))))))))))))))))((((((((....))))).))).....----....................---........----- ( -31.50, z-score = -3.98, R) >droYak2.chr3R 3661579 87 - 28832112 GUUUAUGGUUGCCCGUUGAA-UAGGGCAUCCUUAGGCGCCA--UGCUACUAGGUCUAUAAAAU-----AAAAAUCCAAUACUAUUAC---ACAAUCCA----- (((((.((.(((((......-..))))).)).)))))(((.--........))).........-----...................---........----- ( -17.30, z-score = -0.42, R) >droEre2.scaffold_4770 11097987 87 + 17746568 GCUUGAGGUUGCCCCUUGAA-UCGGGCAGCCGUAGGCGCCA--UGCUACUAAGUGUGUAAAAU-----AAAAAUCCAAUACUAUUAC---ACAAUCCA----- (((((.((((((((......-..)))))))).)))))....--..........(((((((...-----...............))))---))).....----- ( -25.97, z-score = -2.56, R) >droAna3.scaffold_13340 277940 88 + 23697760 -----------UCCUUUGUAUUCGAAAAUCCUCAAGGGC----UCCUAGCACGAGUAGAAAAUAAAAAAAAAACCCAAUACUUUUACGAAAAAAUUCAAAUUG -----------...(((.((((((.....((....))((----.....)).)))))).))).......................................... ( -9.70, z-score = -0.67, R) >consensus GUUUAUGGUUGCCCAAUGGA_UCGGGCAUCCGUAAGCGCCA__UGCUACUAAGUGGGCAAAAU____AAAAAAUCCAAUACUUUUAC___AUAAUCCA_____ (((((.((.(((((.........))))).)).))))).................................................................. (-10.51 = -11.52 + 1.00)

| Location | 14,979,999 – 14,980,101 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 68.59 |
| Shannon entropy | 0.57798 |
| G+C content | 0.42686 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.15 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14979999 102 - 27905053 ----AUAAGUGUUAUGUUUCCUUACCCUAAUCACAGACAUCCAAGU--UUAUGGUUGCCCCAUGGAUCGGGCAUCCGUAAGCGCCA-CUUGCUACUAAGUGGGCAAAAU ----....((((((.((......))..))).)))..........((--((((((.(((((........))))).))))))))((((-((((....))))).)))..... ( -28.90, z-score = -1.74, R) >droSim1.chr3R 21019728 102 + 27517382 ----AUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGU--UUAUGGUUGCCCAAUGGAUCGGGCAUCCGUAAGCGCCA-CUUGCUACUAAGUGGGCGAAAU ----.((((.((......))))))......((.(..........((--((((((.(((((........))))).)))))))).(((-((((....)))))))).))... ( -30.20, z-score = -2.28, R) >droSec1.super_25 47184 102 + 827797 ----AUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGU--UUAUGGUUGCCCAAUGGAUUGGGCAGCCGUAAGCGCUA-CUUGCUACUAAGUGGGCAAAAU ----.((((.((......))))))....................((--((((((((((((((....))))))))))))))))((((-((((....))))).)))..... ( -33.40, z-score = -2.94, R) >droYak2.chr3R 3661606 100 - 28832112 ----AUAAGUGUUACGUUGCCUUAUUUUAAUCACAGGCAUCCAAGU--UUAUGGUUGCCCGUUGAAUAGGGCAUCCUUAGGCGCCA---UGCUACUAGGUCUAUAAAAU ----.............((((((((((.....((.((((.(((...--...))).)))).)).))))))))))..(((((..((..---.))..))))).......... ( -25.90, z-score = -0.99, R) >droEre2.scaffold_4770 11098014 100 + 17746568 ----AUAAGUGUUAUGUUACCUUACCUUAAUCGCAGACAUCCAAGC--UUGAGGUUGCCCCUUGAAUCGGGCAGCCGUAGGCGCCA---UGCUACUAAGUGUGUAAAAU ----.((((.((......))))))........(((.((......((--(((.((((((((........)))))))).)))))((..---.))......)).)))..... ( -27.60, z-score = -1.27, R) >droGri2.scaffold_14906 4708661 109 - 14172833 UUCACUGGUUCGCGUCGUGAUUCAAUGUAGUUAUUCAAUUCACAGUGAGAAAAGAAGACUUUUAAGCUGUGUGUUAAUGAGUGCUGCCAUUUUAGCCUUUUUAAAAACU .....((((((((...))))......(((.(((((.(((.((((((...(((((....)))))..)))))).)))))))).))).)))).................... ( -22.10, z-score = -0.20, R) >consensus ____AUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGU__UUAUGGUUGCCCAAUGAAUCGGGCAUCCGUAAGCGCCA_CUUGCUACUAAGUGGGCAAAAU .......(((..........................................((.(((((........))))).))...((((......)))))))............. (-10.65 = -10.15 + -0.50)
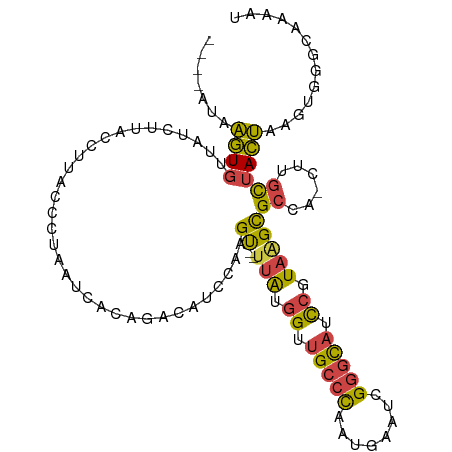
| Location | 14,980,008 – 14,980,127 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.50808 |
| G+C content | 0.43153 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -14.16 |
| Energy contribution | -16.50 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
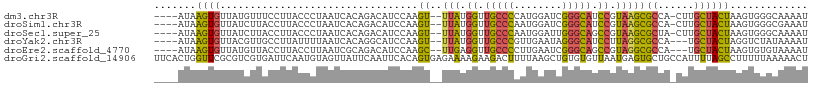
>dm3.chr3R 14980008 119 + 27905053 ACUUAGUAGCAAGUGGCGCUUACGGAUGCCCGAUCCAUGGGGCAACCAUAAACUUGGAUGUCUGUGAUUAGGGUAAGGAAACAUAACACUUAUUUCACAAGAAUUC-GCCAGCUGCUUAA ....((((((...((((......((.(((((........))))).))........((((.(((((((.((((.((.(....).))...))))..)))).)))))))-))))))))))... ( -38.00, z-score = -2.61, R) >droSim1.chr3R 21019737 119 - 27517382 ACUUAGUAGCAAGUGGCGCUUACGGAUGCCCGAUCCAUUGGGCAACCAUAAACUUGGAUGUCUGUGAUUAGGGUAAGGUAAGAUAACACUUAUUUCUCAAGAAUUC-GCCAGCUGCUUAA ....((((((...(((((.((((((((((((((....))))))..(((......)))..))))))))...(((..(((((((......)))))))))).......)-))))))))))... ( -38.60, z-score = -2.64, R) >droSec1.super_25 47193 115 - 827797 ACUUAGUAGCAAGUAGCGCUUACGGCUGCCCAAUCCAUUGGGCAACCAUAAACUUGGAUGUCUGUGAUUAGGGUAAGGUAAGAUAACACUUAUUUCUCAAGAAUUC-G----CUGCUUAA ..........(((((((((((((...(((((((((...((((((.(((......))).)))))).)))).)))))..)))))...........(((....)))..)-)----)))))).. ( -33.30, z-score = -1.85, R) >droYak2.chr3R 3661615 117 + 28832112 ACCUAGUAGCA--UGGCGCCUAAGGAUGCCCUAUUCAACGGGCAACCAUAAACUUGGAUGCCUGUGAUUAAAAUAAGGCAACGUAACACUUAUUUCUCAAGAACUC-UCCAGCUGCUUCA ....((((((.--(((.((((.(((....)))..(((.((((((.(((......))).)))))))))........))))..............(((....)))...-.)))))))))... ( -34.10, z-score = -2.71, R) >droEre2.scaffold_4770 11098023 117 - 17746568 ACUUAGUAGCA--UGGCGCCUACGGCUGCCCGAUUCAAGGGGCAACCUCAAGCUUGGAUGUCUGCGAUUAAGGUAAGGUAACAUAACACUUAUUUCUCAAGAACUC-GCCAGCUGCUUCA ....((((((.--((((((....((.(((((........))))).))....))...((..(((..((...(((((((((......)).))))))).)).)))..))-))))))))))... ( -35.20, z-score = -1.16, R) >droGri2.scaffold_14906 4708688 109 + 14172833 ACUCAUUAACA--CACAGCUUAAAAGUCUUCUUUUC------UCACUGUGAA-UUGAAUAACUAC-AUUGAAUCACGACG-CGAACCAGUGAAUUCAAAUGAACCAAGCUAUCAAUGUGA .....((((..--(((((...(((((....))))).------...)))))..-)))).....(((-(((((.((((....-.......)))).(((....)))........)))))))). ( -18.60, z-score = -0.93, R) >consensus ACUUAGUAGCA__UGGCGCUUACGGAUGCCCGAUCCAUGGGGCAACCAUAAACUUGGAUGUCUGUGAUUAAGGUAAGGUAACAUAACACUUAUUUCUCAAGAACUC_GCCAGCUGCUUAA ....((((((...((((......((.(((((........))))).)).......................(((((((...........)))))))............))))))))))... (-14.16 = -16.50 + 2.34)

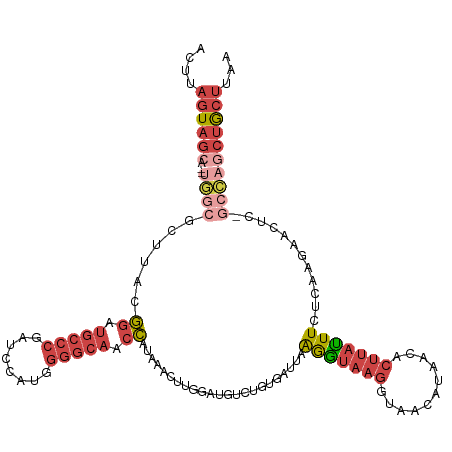
| Location | 14,980,008 – 14,980,127 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.50808 |
| G+C content | 0.43153 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -16.46 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14980008 119 - 27905053 UUAAGCAGCUGGC-GAAUUCUUGUGAAAUAAGUGUUAUGUUUCCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCCAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGU ...(((((.((((-((..(((.((((.....(((...........))).....)))))))..))...((((((((.(((((........))))).)))))))))))).)))))....... ( -37.30, z-score = -3.03, R) >droSim1.chr3R 21019737 119 + 27517382 UUAAGCAGCUGGC-GAAUUCUUGAGAAAUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCAAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGU ...(((((.((((-((..((((((....((((.((......))))))......))).)))..))...((((((((.(((((........))))).)))))))))))).)))))....... ( -37.60, z-score = -3.47, R) >droSec1.super_25 47193 115 + 827797 UUAAGCAG----C-GAAUUCUUGAGAAAUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCAAUGGAUUGGGCAGCCGUAAGCGCUACUUGCUACUAAGU .((((.((----(-((..((((((....((((.((......))))))......))).)))..))...((((((((((((((((....))))))))))))))))))).))))......... ( -36.30, z-score = -3.56, R) >droYak2.chr3R 3661615 117 - 28832112 UGAAGCAGCUGGA-GAGUUCUUGAGAAAUAAGUGUUACGUUGCCUUAUUUUAAUCACAGGCAUCCAAGUUUAUGGUUGCCCGUUGAAUAGGGCAUCCUUAGGCGCCA--UGCUACUAGGU ...((((((((((-..(((..(((((((((((.((......))))))))))..)))..))).)))).(((((.((.(((((........))))).)).)))))))..--))))....... ( -35.80, z-score = -1.48, R) >droEre2.scaffold_4770 11098023 117 + 17746568 UGAAGCAGCUGGC-GAGUUCUUGAGAAAUAAGUGUUAUGUUACCUUACCUUAAUCGCAGACAUCCAAGCUUGAGGUUGCCCCUUGAAUCGGGCAGCCGUAGGCGCCA--UGCUACUAAGU ...((.(((((((-..((..(((((...((((.((......)))))).)))))..))..........(((((.((((((((........)))))))).)))))))))--.))).)).... ( -38.40, z-score = -1.92, R) >droGri2.scaffold_14906 4708688 109 - 14172833 UCACAUUGAUAGCUUGGUUCAUUUGAAUUCACUGGUUCG-CGUCGUGAUUCAAU-GUAGUUAUUCAA-UUCACAGUGA------GAAAAGAAGACUUUUAAGCUGUG--UGUUAAUGAGU ...((((((((...........((((((.((.((..(((-(...))))..)).)-).....))))))-..((((((..------.(((((....)))))..))))))--))))))))... ( -24.20, z-score = -0.23, R) >consensus UUAAGCAGCUGGC_GAAUUCUUGAGAAAUAAGUGUUAUGUUACCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCAAUGAAUCGGGCAUCCGUAAGCGCCA__UGCUACUAAGU ...((((..((((....(((....)))........................................(((((.((.(((((........))))).)).)))))))))..))))....... (-16.46 = -17.30 + 0.84)

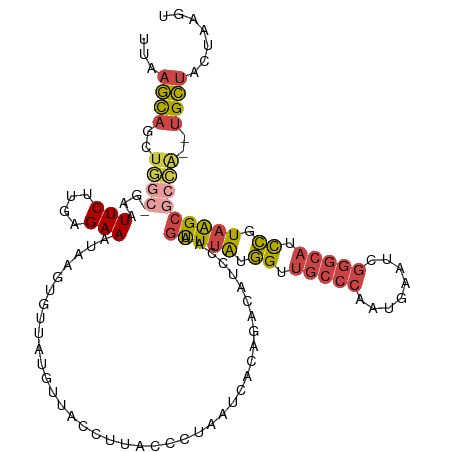
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:03 2011