
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,958,425 – 14,958,525 |
| Length | 100 |
| Max. P | 0.881839 |

| Location | 14,958,425 – 14,958,525 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 54.57 |
| Shannon entropy | 0.88727 |
| G+C content | 0.36964 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -3.47 |
| Energy contribution | -3.20 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.93 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.15 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
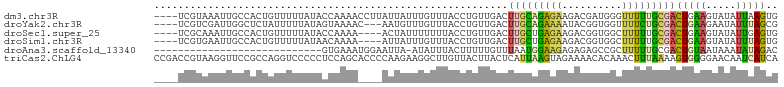
>dm3.chr3R 14958425 100 - 27905053 ----UCGUAAAUUGCCACUGUUUUUAUACCAAAACCUUAUUAUUUGUUUACCUGUUGACUUGCAGAGAAGACGAUGGGUUUUUGCGACUGAAGUAUAUUAAGUG ----(((((((..(((...(((((......)))))........((((((.(((((......)))).).))))))..))).)))))))................. ( -18.30, z-score = -0.74, R) >droYak2.chr3R 3641740 97 - 28832112 ----UCGUCGAUUGGCUCUAUUUUUAUAGUAAAAC---AAUGUUUGUUUACCUGUUGACUUGCAGAAAAUACGGUGGUUUUCUGCGACUGAAGAAUAUUUAGCG ----..((((((.((..((((....))))..((((---(.....))))).)).))))))(((((((((((......)))))))))))(((((.....))))).. ( -24.20, z-score = -2.16, R) >droSec1.super_25 27627 96 + 827797 ----UCGCAAAUUGCCACUGUUUUUAUACCAAAA----ACUAUUUUUUUACCUGUUGACUUGCUGAGAAGACGGUGGCUUUUUGCGACUGAAGUAUAUUGAGUG ----.((((((..((((((((((((.((.(((.(----((.............)))...))).)).))))))))))))..))))))(((.((.....)).))). ( -29.82, z-score = -4.97, R) >droSim1.chr3R 21000050 96 + 27517382 ----UCGUGAAUUGCCACUGUUUUUAUAACAAAA----AUUAUUUGUUUACCUGUUGACUUGCUGAGAAGACGGUGGCUUUUUGCGACUGAAGUAUAUUUAGUG ----.((..((..((((((((((((..((((((.----....)))))).....((......))...))))))))))))..))..))((((((.....)))))). ( -29.20, z-score = -4.71, R) >droAna3.scaffold_13340 256829 75 + 23697760 ----------------------------GUGAAAUGGAAUUA-AUAUUUACUUUUUGUUUAAUGGAAGAGAGAGCCGCUUUUUGCGACUGUAAUAAAUAUAGAC ----------------------------..............-(((((((((((((.(((....))).)))))).(((.....))).......))))))).... ( -10.80, z-score = -0.64, R) >triCas2.ChLG4 4690747 104 + 13894384 CCGACCGUAAGGUUCCGCCAGGUCCCCCUCCAGCACCCCAAGAAGGCUUGUUACUUACUCAUUAAGUAGAAAACACAAACUUUAAAAGUGGGGAACAAUCAUCA ..((((....))))..((.(((....)))...)).(((((..((((.((((((((((.....))))))......)))).)))).....)))))........... ( -23.50, z-score = -0.99, R) >consensus ____UCGUAAAUUGCCACUGUUUUUAUACCAAAA____AUUAUUUGUUUACCUGUUGACUUGCAGAGAAGACGGUGGCUUUUUGCGACUGAAGUAUAUUUAGUG ...........................................................(((((((((..........)))))))))(((((.....))))).. ( -3.47 = -3.20 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:59 2011