| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,954,973 – 14,955,074 |
| Length | 101 |
| Max. P | 0.794658 |

| Location | 14,954,973 – 14,955,074 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.54633 |
| G+C content | 0.53154 |
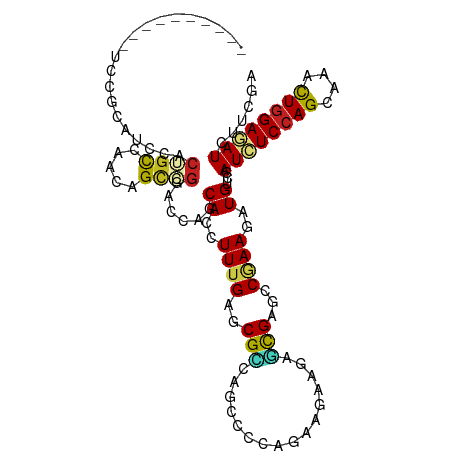
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.08 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
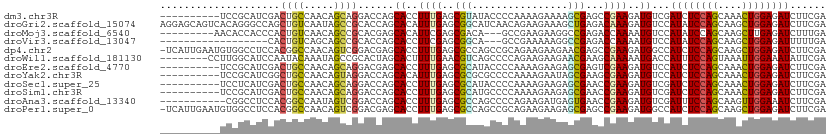
>dm3.chr3R 14954973 101 + 27905053 ----------UCCGCAUCGACUGCCAACAGCAGGACCAGCACCUUUGAGCGUAUACCCCAAAAGAAAAGCGAGCCGAAGAUGUCGAUCUCCAGCAAACUGGAGAUCUUCGA ----------......((((((((.....)))).....(((.(((((.((......................))))))).))).(((((((((....))))))))).)))) ( -28.75, z-score = -2.42, R) >droGri2.scaffold_15074 7129427 111 + 7742996 AGGAGCAGUCACAGGGCCAGCUGUCAAUAGCCGCACCAGCACAUUUGAGCGGCAUCAACAGAAGAAAGCUGAGACAAAGAUGUCCAUAUCCAGCAAGCUGGAGAUCUUCGA .((((..(((...(((((((((.((....(((((..(((.....))).))))).((....)).)).)))))..........))))...(((((....)))))))))))).. ( -32.30, z-score = -1.19, R) >droMoj3.scaffold_6540 14215776 99 + 34148556 ---------AACACCACCCACUGUCAACAGCCGCACGAGCACAUUCGAGCGACA---GCCGAAGAAGGCCGAGACCAAAAUGUCCAUAUCCAGCAAGCUUGAGAUCUUUGA ---------...........(((....)))..((....))...(((((((....---(((......))).(.(((......))))...........)))))))........ ( -17.80, z-score = -0.11, R) >droVir3.scaffold_13047 1769814 90 - 19223366 ------------------CACUGUCAGCAGCCGCACCAGCACCUUCGAGCGGCA---GCCGAAAAAGGCCGAGACCAAAAUGUCCAUAUCCAGCAAGCUGGAGAUUUUUGA ------------------....(((....(((((..............))))).---(((......)))...)))(((((.(((....(((((....))))))))))))). ( -25.64, z-score = -1.30, R) >dp4.chr2 6895242 110 + 30794189 -UCAUUGAAUGUGGCCUCCACGGCCAACAGUCGGACGAGCACCUUUGAGCGCCAGCCGCAGAAGAAGAACGAGCCGAAGAUGGCCAUCUCCAGCAAGCUGGAGAUCUUCGA -...(((((.((((...))))(((((....((((.((..(..((((..(((.....))).))))..)..))..))))...)))))((((((((....)))))))).))))) ( -39.00, z-score = -1.88, R) >droWil1.scaffold_181130 1854471 103 - 16660200 --------CCUUGGCAUCCAAUACAAAUAGCCGCACUAGCACUUUUGAACGUCAGCCCCAGAAGAAGAACGAAGCAAAAAUGACCAUUUCCAGUAAAUUGGAAAUAUUCGA --------....(((..............)))((....)).((((((...........))))))................(((..((((((((....))))))))..))). ( -15.74, z-score = -0.25, R) >droEre2.scaffold_4770 11074821 101 - 17746568 ----------UCCGCAUCGACUGCCAACAGCAGGACGAGCACCUUUGAGCGCAUACCCCAAAAGAAGAGCGAGUCGAAGAUGUCCAUCUCCAGCAAACUGGAGAUCUUCGA ----------..(((.(((.((((.....))))..)))((.(......).))................)))..(((((((......(((((((....)))))))))))))) ( -30.00, z-score = -2.11, R) >droYak2.chr3R 3638292 101 + 28832112 ----------UCCGCAUCGGCUGCCAACAGUAGGACCAGCACAUUUGAGCGCGCGCCCCAAAAGAAUAGCGAAGCGAAGAUGUCCAUCUCCAGCAAACUGGAGAUCUUCGA ----------..(((....((((((.......))..))))...((((.((....))..))))......)))...((((((......(((((((....))))))))))))). ( -28.50, z-score = -0.92, R) >droSec1.super_25 24216 101 - 827797 ----------UCCUCAUCGACUGCCAACAGCAGGACCAGCACCUUUGAGCGCAUACCCCAAAAGAAGAGCGAACCGAAGAUGUCGAUCUCCAGCAAACUGGAGAUCUUCGA ----------......((((((((.....)))).....(((.(((((..(((................)))...))))).))).(((((((((....))))))))).)))) ( -30.09, z-score = -2.93, R) >droSim1.chr3R 20996632 101 - 27517382 ----------UCCGCAUCGACUGCCAACAGCAGGACCAGCACCUUUGAGCGCAUGCCCCAAAAGAAGAGCGAACCGAAGAUGUCGAUCUCCAGCAAACUGGAGAUCUUCGA ----------......((((((((.....)))).....(((.(((((..(((................)))...))))).))).(((((((((....))))))))).)))) ( -30.09, z-score = -2.21, R) >droAna3.scaffold_13340 253514 100 - 23697760 -----------CGGCCUCCACGGCCAAUAGUCGGACCAGCACCUUUGAGCGCCAGCCCCAGAAGAUGAGUGAACCGAAGAUGUCGAUUUCCAGCAAGUUGGAAAUCUUCGA -----------.((((.....)))).((..((((..((.((.((((..((....))....)))).))..))..))))..))...(((((((((....)))))))))..... ( -30.10, z-score = -1.62, R) >droPer1.super_0 751652 110 + 11822988 -UCAUUGAAUGUGGCCUCCACGGCCAACAGUCGGACGAGCACCUUUGAGCGCCAGCCGCAGAAGAAGAGCGAGCCGAAGAUGGCCAUCUCCAGCAAGCUGGAGAUCUUCGA -...(((((.((((...))))(((((....((((.((..(..((((..(((.....))).))))..)..))..))))...)))))((((((((....)))))))).))))) ( -40.40, z-score = -1.83, R) >consensus __________UCCGCAUCCACUGCCAACAGCCGGACCAGCACCUUUGAGCGCCAGCCCCAGAAGAAGAGCGAGCCGAAGAUGUCCAUCUCCAGCAAACUGGAGAUCUUCGA ....................((((.....))))......((..((((..(((................)))...))))..))...((((((((....))))))))...... (-13.44 = -13.08 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:58 2011