| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,952,125 – 7,952,216 |
| Length | 91 |
| Max. P | 0.519989 |

| Location | 7,952,125 – 7,952,216 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Shannon entropy | 0.37517 |
| G+C content | 0.45106 |
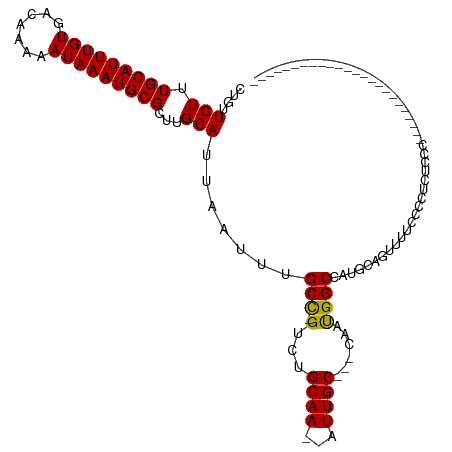
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -14.10 |
| Energy contribution | -13.70 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
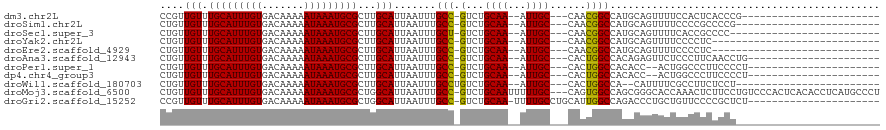
>dm3.chr2L 7952125 91 - 23011544 CCGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CAACGGCCAUGCAGUUUUCCACUCACCCG----------------------- ((((((...(((((((((((..((((.(((((....))))).))))...-))).))))--).)))---)))))).......................----------------------- ( -21.80, z-score = -1.21, R) >droSim1.chr2L 7747697 90 - 22036055 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CAACGGCCAUGCAGUUUUCCCCGCCCCG------------------------ (((((((.(((((((((.......)))))))))...))).......(((-((..((..--...))---..)))))...))))..............------------------------ ( -21.80, z-score = -1.26, R) >droSec1.super_3 3465877 89 - 7220098 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCU-GUCUGCAA--AUUGC---CAACGGCCAUGCAGUUUUCACCGCCCC------------------------- ..((((((.((((((((((((.((((.(((((....))))).))))..)-))).))))--).)))---.))))))...((.((....)).))...------------------------- ( -23.30, z-score = -1.36, R) >droYak2.chr2L 17383277 86 + 22324452 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CAACGGCCAUGCAGUUUUCCCCUC---------------------------- (((((((.(((((((((.......)))))))))...))).......(((-((..((..--...))---..)))))...))))..........---------------------------- ( -21.80, z-score = -1.70, R) >droEre2.scaffold_4929 16874645 86 - 26641161 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CAACGGCCAUGCAGUUUUCCCCUC---------------------------- (((((((.(((((((((.......)))))))))...))).......(((-((..((..--...))---..)))))...))))..........---------------------------- ( -21.80, z-score = -1.70, R) >droAna3.scaffold_12943 3682827 92 + 5039921 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CACUGGCCACAGAGUUCUCCCUUCAACCUG---------------------- ((((.(((.(((((((((((..((((.(((((....))))).))))...-))).))))--).)))---....))).))))..................---------------------- ( -19.80, z-score = -0.99, R) >droPer1.super_1 3472404 90 + 10282868 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CACUGGCCACACC--ACUGGCCCUUCCCCU---------------------- ....(((.(((((((((.......)))))))))...)))..(((((((.-....))))--)))((---((.(((.....))--).)))).........---------------------- ( -22.90, z-score = -1.96, R) >dp4.chr4_group3 6373420 90 + 11692001 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC-GUCUGCAA--AUUGC---CACUGGCCACACC--ACUGGCCCUUCCCCU---------------------- ....(((.(((((((((.......)))))))))...)))..(((((((.-....))))--)))((---((.(((.....))--).)))).........---------------------- ( -22.90, z-score = -1.96, R) >droWil1.scaffold_180703 582383 89 + 3946847 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCUGUCUGCAA--AUUGC---CACUGGCCA--CAUUUUCGCCUUCUCCU------------------------ ..((.(((((((.....((((.((((.(((((....))))).))))...)))))))))--)).))---....(((..--.......))).......------------------------ ( -18.20, z-score = -1.01, R) >droMoj3.scaffold_6500 29752130 116 + 32352404 CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUGGCAUUAAUUUGCC-GUCUGCAAUUUUUGC---CAGUGGCCAGCGGGCACCAAACUCUUCCUGUCCCACUCACACCUCAUGCCCU ..((((...((((((((.......))))))))(((((((..((((.((.-....))))))..)))---))))...))))(((((..............................))))). ( -32.21, z-score = -1.55, R) >droGri2.scaffold_15252 4133507 96 + 17193109 CCGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUGGCAUUAAUUUGCC-GUCUGCAA-UUUUGCCUGCAUUGGCCAGACCCUGCUGUUCCCCGCUCU---------------------- .........((((((((.......))))))))((.((((......))))-(((((...-....(((......)))))))).............))...---------------------- ( -24.61, z-score = -1.29, R) >consensus CUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCC_GUCUGCAA__AUUGC___CAAUGGCCAUGCAGUUUUCCCCUCUCCC________________________ (((.((...(((.(((((((..((((.(((((....))))).))))....))).))))....)))...)).))).............................................. (-14.10 = -13.70 + -0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:48 2011