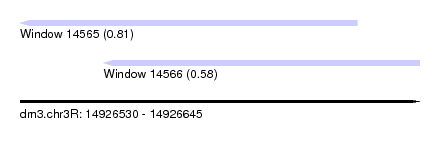
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,926,530 – 14,926,645 |
| Length | 115 |
| Max. P | 0.805407 |

| Location | 14,926,530 – 14,926,627 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.45 |
| Shannon entropy | 0.55820 |
| G+C content | 0.34095 |
| Mean single sequence MFE | -18.28 |
| Consensus MFE | -7.76 |
| Energy contribution | -8.15 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
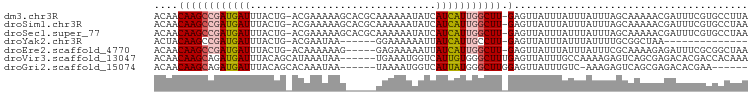
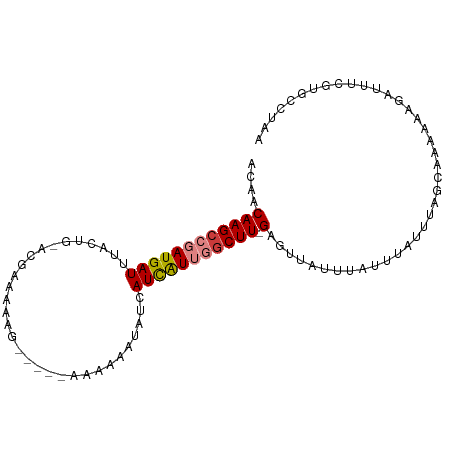
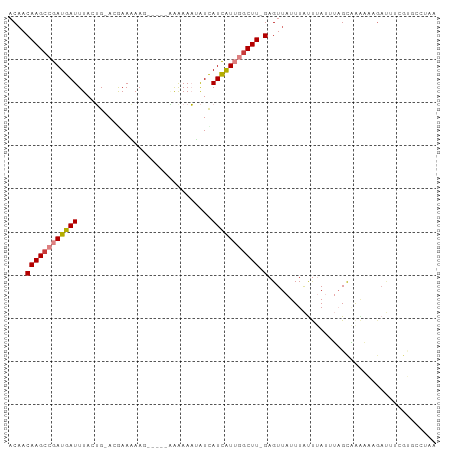
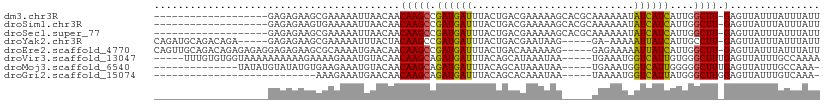
>dm3.chr3R 14926530 97 - 27905053 ACAACAAGCCGAUGAUUUACUG-ACGAAAAAGCACGCAAAAAAUAUCAUCAUUGGCUU-GAGUUAUUUAUUUAUUUAGCAAAAACGAUUUCGUGCCUUA ....((((((((((((....((-.((........)))).........)))))))))))-).((((..........))))....(((....)))...... ( -18.22, z-score = -1.46, R) >droSim1.chr3R 20968241 97 + 27517382 ACAACAAGCCGAUGAUUUACUG-ACGAAAAAGCACGCAAAAAAUAUCAUCAUUGGCUU-GAGUUAUUUAUUUAUUUAGCAAAAACGAUUUCGUGCCUAA ....((((((((((((....((-.((........)))).........)))))))))))-).((((..........))))....(((....)))...... ( -18.22, z-score = -1.52, R) >droSec1.super_77 107558 97 + 117255 ACAACAAGCCGAUGAUUUACUG-ACGAAAAAGCACGCAAAAAAUAUCAUCAUUGGCUU-GAGUUAUUUAUUUAUUUAGCAAAAACGAUUUCGUGCCUAA ....((((((((((((....((-.((........)))).........)))))))))))-).((((..........))))....(((....)))...... ( -18.22, z-score = -1.52, R) >droYak2.chr3R 3610285 77 - 28832112 ACUACAAGCCGAUGAUUUACUG-ACGAAUAA------GGAAAAAAUUAUCAUUGCCUU-GAGUUAUUUAUUUAUUUUGCGGCUAA-------------- ......((((((((((((.((.-.......)------)....))))))))...((..(-((((.....)))))....))))))..-------------- ( -14.00, z-score = -1.76, R) >droEre2.scaffold_4770 11042753 92 + 17746568 ACAACAAGCCGAUGAUUUACUG-ACAAAAAAG-----GAGAAAAAUUAUCAUUGGCUU-GAGUUAUUUAUUUAUUUCGCAAAAGAGAUUUCGCGGCUAA ....((((((((((((......-.........-----..........)))))))))))-)...............((((.(((....))).)))).... ( -19.81, z-score = -2.04, R) >droVir3.scaffold_13047 1734302 93 + 19223366 ACAACAAGCAGAUGAUUUACAGCAUAAAUAA------UGAAAUGGUCAUUGUGGGCUUUGAGUUAUUUGCCAAAAGAGUCAGCGAGACACGACCACAAA .......((((((((((((.(((....((((------(((.....)))))))..))).)))))))))))).......(((.....)))........... ( -24.40, z-score = -3.00, R) >droGri2.scaffold_15074 7099561 86 - 7742996 ACAACAAGCAGAUGAUUUACAGCACAAAUAA------UAAAAUGGUCAUUAUGGGCUUGGAGUUAUUUGUC-AAAGAGUCAGCGAGACACGAA------ .......(((((((((((..(((....((((------(.........)))))..)))..))))))))))).-.....(((.....))).....------ ( -15.10, z-score = -0.77, R) >consensus ACAACAAGCCGAUGAUUUACUG_ACGAAAAAG_____AAAAAAUAUCAUCAUUGGCUU_GAGUUAUUUAUUUAUUUAGCAAAAAAGAUUUCGUGCCUAA .....(((((((((((...............................)))))))))))......................................... ( -7.76 = -8.15 + 0.39)
| Location | 14,926,554 – 14,926,645 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.61 |
| Shannon entropy | 0.62653 |
| G+C content | 0.31801 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -4.71 |
| Energy contribution | -4.18 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
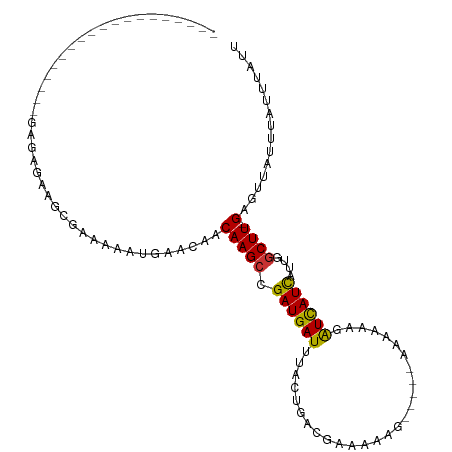
>dm3.chr3R 14926554 91 - 27905053 -------------------GAGAGAAGCGAAAAAUUAACAACAAGCCGAUGAUUUACUGACGAAAAAGCACGCAAAAAAUAUCAUCAUUGGCUU-GAGUUAUUUAUUUAUU -------------------.........(((.((.((((..((((((((((((....((.((........)))).........)))))))))))-).)))).)).)))... ( -16.72, z-score = -2.15, R) >droSim1.chr3R 20968265 91 + 27517382 -------------------GAGAGAAGUGAAAAAUUAACAACAAGCCGAUGAUUUACUGACGAAAAAGCACGCAAAAAAUAUCAUCAUUGGCUU-GAGUUAUUUAUUUAUU -------------------......((((((.((.((((..((((((((((((....((.((........)))).........)))))))))))-).)))).)).)))))) ( -19.02, z-score = -3.08, R) >droSec1.super_77 107582 91 + 117255 -------------------GAGAGAAGCGAAAAAUUAACAACAAGCCGAUGAUUUACUGACGAAAAAGCACGCAAAAAAUAUCAUCAUUGGCUU-GAGUUAUUUAUUUAUU -------------------.........(((.((.((((..((((((((((((....((.((........)))).........)))))))))))-).)))).)).)))... ( -16.72, z-score = -2.15, R) >droYak2.chr3R 3610295 99 - 28832112 CAGAUGCAGACAGA-----GAGAGAAGCGAAAAAUUUACUACAAGCCGAUGAUUUACUGACGAAUAAG-----GA-AAAAAUUAUCAUUGCCUU-GAGUUAUUUAUUUAUU ....(((.......-----.......)))..((((..(((.((((.(((((((.......(......)-----..-.......))))))).)))-)))).))))....... ( -11.63, z-score = 0.06, R) >droEre2.scaffold_4770 11042777 105 + 17746568 CAGUUGCAGACAGAGAGAGGAGAGAAGCGCAAAAUGAACAACAAGCCGAUGAUUUACUGACAAAAAAG-----GAGAAAAAUUAUCAUUGGCUU-GAGUUAUUUAUUUAUU ....(((.....................)))((((((....((((((((((((...............-----..........)))))))))))-)..))))))....... ( -17.81, z-score = -1.32, R) >droVir3.scaffold_13047 1734326 101 + 19223366 -----UUUGUGUGGUAAAAAAAAAAGAAAAGAAAUGUACAACAAGCAGAUGAUUUACAGCAUAAAUAA-----UGAAAUGGUCAUUGUGGGCUUUGAGUUAUUUGCCAAAA -----.((((...(((....................))).))))((((((((((((.(((....((((-----(((.....)))))))..))).))))))))))))..... ( -23.05, z-score = -2.81, R) >droMoj3.scaffold_6540 14185831 91 - 34148556 --------------UAUAUGUAUAUGUGAAGAAAUGUACAACAAGCAGAUGAUUUACAGCAUAAAUAA-----UGAAAUGGUCAUUGGGGGCUUUGAGUUAUUUGCCAAA- --------------....(((((((........)))))))....((((((((((((.(((.....(((-----(((.....))))))...))).))))))))))))....- ( -25.80, z-score = -3.89, R) >droGri2.scaffold_15074 7099579 78 - 7742996 ---------------------------AAAGAAAUGAACAACAAGCAGAUGAUUUACAGCACAAAUAA-----UAAAAUGGUCAUUAUGGGCUUGGAGUUAUUUGUCAAA- ---------------------------...((....(((..(((((.(((((((..............-----......)))))))....)))))..))).....))...- ( -13.55, z-score = -1.38, R) >consensus ___________________GAGAGAAGCGAAAAAUGAACAACAAGCCGAUGAUUUACUGACGAAAAAG_____AAAAAAGAUCAUCAUUGGCUU_GAGUUAUUUAUUUAUU ..........................................((((.((((((...........................))))))....))))................. ( -4.71 = -4.18 + -0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:53 2011