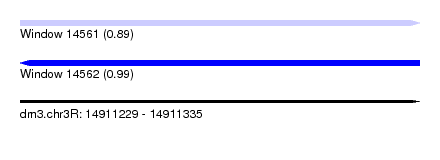
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,911,229 – 14,911,335 |
| Length | 106 |
| Max. P | 0.991080 |

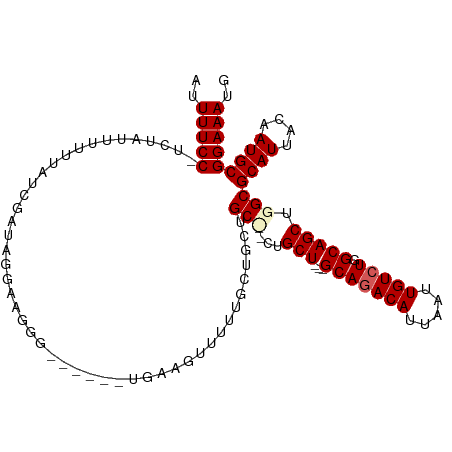
| Location | 14,911,229 – 14,911,335 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Shannon entropy | 0.31940 |
| G+C content | 0.43117 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14911229 106 + 27905053 AUUUUCCUUCUAUUUUUUAUCGAUAUGAAGGGAAGGGGUGAAGUUUUUUCUACUGC---UGCU---GCAGACAUUAAUUGUCUGGCAGCUGGCGCAUUACAAUGCGGAAAUG .(((((((((((((.......)))).)))))))))...........(((((...((---((((---(((((((.....))))).))))).)))((((....))))))))).. ( -34.00, z-score = -3.32, R) >droSim1.chr3R 20951440 94 - 27517382 AUUUUCCCUCUAUUUUUUAUCGAUAGGAAGGG------UGAAGUUUU------UGC---CGCU---GCAGACAUUAAUUGUCUGGCAGCUGGCGCAUUACAAUGCGGAAAUG .(((.(((((((((.......)))))..))))------.)))(((((------(((---((((---(((((((.....))))).))))).)))((((....)))))))))). ( -32.40, z-score = -3.40, R) >droSec1.super_77 89459 103 - 117255 AUUUUCCCUCUAUUUUUUAUCGAUAGGAAGGG------UGAAGUUUUUGCUGCUGCCGCUGCU---GCAGACAUUAAUUGUCUGGCAGCUGGCGCAUUACAAUGCGGAAAUG .(((.(((((((((.......)))))..))))------.)))(((((((((((.((((((((.---.((((((.....))))))))))).)))))).......)))))))). ( -36.31, z-score = -2.98, R) >droYak2.chr3R 3595180 103 + 28832112 AUUUUCC-----UUUUUUAUAGAUAGGGUGAA----GUUUUCGCUGCUGCUUCUGCUUCUGCUGCUGCACACAUUAAUUGUCUGGCAGCUGGCGCAUUACAAUGCGGAAAUG ..(((((-----.................(((----((....((....))....))))).((((((((..(((.....)))...))))).)))((((....))))))))).. ( -27.90, z-score = -0.67, R) >droEre2.scaffold_4770 11027532 102 - 17746568 AUUUUCC-----UUUUUCAUCGAUAGGAAGGG-----GUGAAGUUUUUGUUGCUGCUGCUGCUUCUGCAGACAUUAAUUGUCUGGCAGCUGGCGCAUUACAAUGCGGAAAUG ..(((((-----...((((((..........)-----)))))((..(((((((.((((((((.....((((((.....))))))))))).))))))..)))).))))))).. ( -32.30, z-score = -1.82, R) >consensus AUUUUCC_UCUAUUUUUUAUCGAUAGGAAGGG______UGAAGUUUUUGCUGCUGC__CUGCU___GCAGACAUUAAUUGUCUGGCAGCUGGCGCAUUACAAUGCGGAAAUG ..(((((...............................................(((...(((....((((((.....))))))..))).)))((((....))))))))).. (-18.52 = -19.04 + 0.52)

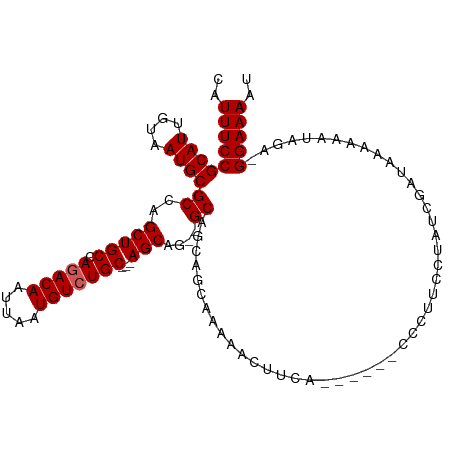
| Location | 14,911,229 – 14,911,335 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Shannon entropy | 0.31940 |
| G+C content | 0.43117 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -18.38 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14911229 106 - 27905053 CAUUUCCGCAUUGUAAUGCGCCAGCUGCCAGACAAUUAAUGUCUGC---AGCA---GCAGUAGAAAAAACUUCACCCCUUCCCUUCAUAUCGAUAAAAAAUAGAAGGAAAAU ..(((((((((....)))))...(((((((((((.....)))))).---.)))---))....))))...............(((((.(((.........))))))))..... ( -25.90, z-score = -3.60, R) >droSim1.chr3R 20951440 94 + 27517382 CAUUUCCGCAUUGUAAUGCGCCAGCUGCCAGACAAUUAAUGUCUGC---AGCG---GCA------AAAACUUCA------CCCUUCCUAUCGAUAAAAAAUAGAGGGAAAAU .......((((....))))(((.(((((.(((((.....)))))))---))))---)).------.........------((((..((((.........))))))))..... ( -28.70, z-score = -4.02, R) >droSec1.super_77 89459 103 + 117255 CAUUUCCGCAUUGUAAUGCGCCAGCUGCCAGACAAUUAAUGUCUGC---AGCAGCGGCAGCAGCAAAAACUUCA------CCCUUCCUAUCGAUAAAAAAUAGAGGGAAAAU ..........((((..((((((.(((((((((((.....)))))).---.)))))))).)))))))........------((((..((((.........))))))))..... ( -30.80, z-score = -3.08, R) >droYak2.chr3R 3595180 103 - 28832112 CAUUUCCGCAUUGUAAUGCGCCAGCUGCCAGACAAUUAAUGUGUGCAGCAGCAGAAGCAGAAGCAGCAGCGAAAAC----UUCACCCUAUCUAUAAAAAA-----GGAAAAU ..(((((((((....))))((..(((((.(.(((.....))).)))))).)).((((.....((....)).....)----))).................-----))))).. ( -25.50, z-score = -1.48, R) >droEre2.scaffold_4770 11027532 102 + 17746568 CAUUUCCGCAUUGUAAUGCGCCAGCUGCCAGACAAUUAAUGUCUGCAGAAGCAGCAGCAGCAACAAAAACUUCAC-----CCCUUCCUAUCGAUGAAAAA-----GGAAAAU ..(((((((((....))))((..(((((((((((.....)))))).....))))).)).................-----....................-----))))).. ( -24.60, z-score = -2.16, R) >consensus CAUUUCCGCAUUGUAAUGCGCCAGCUGCCAGACAAUUAAUGUCUGC___AGCAG__GCAGCAGCAAAAACUUCA______CCCUUCCUAUCGAUAAAAAAUAGA_GGAAAAU ..(((((((((....))))((...((((((((((.....)))))).....))))..))...............................................))))).. (-18.38 = -18.82 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:49 2011