
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,895,400 – 14,895,505 |
| Length | 105 |
| Max. P | 0.997580 |

| Location | 14,895,400 – 14,895,505 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.56 |
| Shannon entropy | 0.60449 |
| G+C content | 0.55910 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -15.62 |
| Energy contribution | -17.90 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
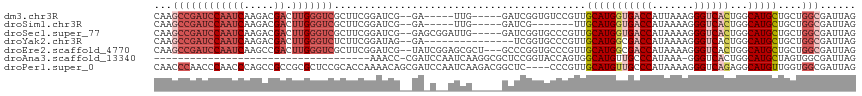
>dm3.chr3R 14895400 105 + 27905053 CAAGCCGAUCCAAUCAAGACGACUUGGGUCGCUUCGGAUCG--GA-----UUG-----GAUCGGUGUCCGUUGCAUGGUGACCAUUAAAGGGUCACUGGCAUGCUGCUGGCGAUUAG ...((((((((((((....((((....))))(....)....--))-----)))-----)))))))(((.((.(((((((((((.......))))))...))))).)).)))...... ( -46.00, z-score = -3.91, R) >droSim1.chr3R 20935916 98 - 27517382 CAAGCCGAUCCAAUCAAGACGACUUGGGUCGCUUCGGAUCG--GA-----UUG-----GAUCG-------UUGCAUGGUGACCAUAAAAGGGUCACUGGCAUGCUGCUGGCGAUUAG ...((((((((((((....((((....))))(....)....--))-----)))-----))))(-------(.(((((((((((.......))))))...))))).)).)))...... ( -41.20, z-score = -3.91, R) >droSec1.super_77 73978 110 - 117255 CAAGCCGAUCCAAUCAAGACGACUUGGGUCGCUUCGGAUCG--GAGCGGAUUG-----GAUCGGUGCCCGUUGCAUGGUGACCAUAAAAGGGUCACUGGCAUGCUGCUGGCGAUUAG ...((((((((((((.....(((....)))((((((...))--)))).)))))-----)))))))(((.((.(((((((((((.......))))))...))))).)).)))...... ( -53.90, z-score = -5.15, R) >droYak2.chr3R 3578860 100 + 28832112 CAAGCCGAUCCAAUCAAGACGACUUGGGUCUCUUCGGAUAG--GA---------------UCGGUGCCCGUUGCAUGGCGACCAUAAAAGGGUCACUGGCAUGCUGCUGGCGAUUAG ...((((((((.(((.(((.(((....))))))...))).)--))---------------)))))(((.((.(((((.(((((.......))))...).))))).)).)))...... ( -42.00, z-score = -3.40, R) >droEre2.scaffold_4770 11012134 112 - 17746568 CAAGCCGAUCCAAUCAAGCCGACUUGGGUCGCUUCGGAUCG--UAUCGGAGCGCU---GCCCGGUGCCCGUUGCAUGGCGACCAUAAAAGGGUCACUGGCAUGCUGCUGGCGAUUAG ...((((((((((((.....)).)))))))(((((((....--..)))))))(((---(((.(((((((.((..((((...)))).)).))).)))))))).))....)))...... ( -43.80, z-score = -1.05, R) >droAna3.scaffold_13340 17530618 79 + 23697760 ------------------------------------AAACC-CGAUCCAAUCAAGGCGCUCCGGUACCAGUGGCAUGUUGCCCAUAAA-GGGUCACUGGCAUGCUAGUGGCGAUUAG ------------------------------------..(((-.((.((......))...)).))).(((.(((((((((((((.....-))))....))))))))).)))....... ( -25.90, z-score = -0.99, R) >droPer1.super_0 665680 113 + 11822988 CAACCCAACCCAACCCAGCCGCCGCGCUCCGCACCAAAACAGCGAUCCAAUCAAGACGGCUC----CCCGUUGCAUGUUGCCCAUAAAAGGGUCAGAGGCAUGUUGGUGGCGAUUAG .................((((((.((((............))))..........(((((...----.)))))(((((((((((......))))....))))))).))))))...... ( -31.10, z-score = -0.34, R) >consensus CAAGCCGAUCCAAUCAAGACGACUUGGGUCGCUUCGGAUCG__GA_C__AUCG_____GAUCGGUGCCCGUUGCAUGGUGACCAUAAAAGGGUCACUGGCAUGCUGCUGGCGAUUAG ...((((((((((((.....)).)))))))..........................................(((((((((((.......))))))...)))))....)))...... (-15.62 = -17.90 + 2.28)
| Location | 14,895,400 – 14,895,505 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.56 |
| Shannon entropy | 0.60449 |
| G+C content | 0.55910 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -17.61 |
| Energy contribution | -19.96 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
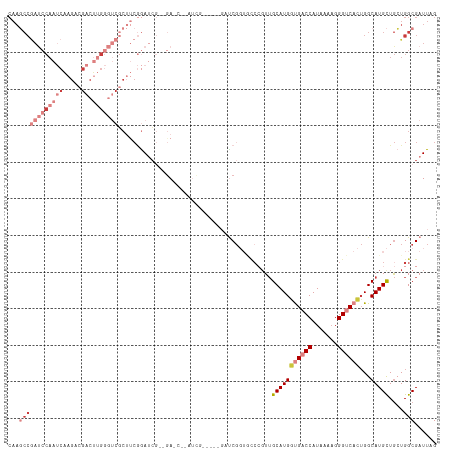
>dm3.chr3R 14895400 105 - 27905053 CUAAUCGCCAGCAGCAUGCCAGUGACCCUUUAAUGGUCACCAUGCAACGGACACCGAUC-----CAA-----UC--CGAUCCGAAGCGACCCAAGUCGUCUUGAUUGGAUCGGCUUG ......(((....(((((...((((((.......)))))))))))..........((((-----(((-----((--......((..((((....))))..))))))))))))))... ( -37.80, z-score = -4.02, R) >droSim1.chr3R 20935916 98 + 27517382 CUAAUCGCCAGCAGCAUGCCAGUGACCCUUUUAUGGUCACCAUGCAA-------CGAUC-----CAA-----UC--CGAUCCGAAGCGACCCAAGUCGUCUUGAUUGGAUCGGCUUG ......(((....(((((...((((((.......)))))))))))..-------.((((-----(((-----((--......((..((((....))))..))))))))))))))... ( -37.80, z-score = -5.00, R) >droSec1.super_77 73978 110 + 117255 CUAAUCGCCAGCAGCAUGCCAGUGACCCUUUUAUGGUCACCAUGCAACGGGCACCGAUC-----CAAUCCGCUC--CGAUCCGAAGCGACCCAAGUCGUCUUGAUUGGAUCGGCUUG ......(((....(((((...((((((.......)))))))))))....))).((((((-----(((((.(((.--((...)).)))(((....))).....))))))))))).... ( -43.70, z-score = -4.47, R) >droYak2.chr3R 3578860 100 - 28832112 CUAAUCGCCAGCAGCAUGCCAGUGACCCUUUUAUGGUCGCCAUGCAACGGGCACCGA---------------UC--CUAUCCGAAGAGACCCAAGUCGUCUUGAUUGGAUCGGCUUG ......(((....(((((...((((((.......)))))))))))....))).((((---------------((--(.(((..(((((((....))).))))))).))))))).... ( -38.30, z-score = -3.60, R) >droEre2.scaffold_4770 11012134 112 + 17746568 CUAAUCGCCAGCAGCAUGCCAGUGACCCUUUUAUGGUCGCCAUGCAACGGGCACCGGGC---AGCGCUCCGAUA--CGAUCCGAAGCGACCCAAGUCGGCUUGAUUGGAUCGGCUUG ......((((((.((.((((.(((.(((((.(((((...))))).)).))))))..)))---))))))......--.(((((((((((((....))).)))...))))))))))... ( -42.10, z-score = -1.45, R) >droAna3.scaffold_13340 17530618 79 - 23697760 CUAAUCGCCACUAGCAUGCCAGUGACCC-UUUAUGGGCAACAUGCCACUGGUACCGGAGCGCCUUGAUUGGAUCG-GGUUU------------------------------------ ......((((...(((((....((.(((-.....))))).)))))...))))(((.((...((......)).)).-)))..------------------------------------ ( -24.00, z-score = -0.44, R) >droPer1.super_0 665680 113 - 11822988 CUAAUCGCCACCAACAUGCCUCUGACCCUUUUAUGGGCAACAUGCAACGGG----GAGCCGUCUUGAUUGGAUCGCUGUUUUGGUGCGGAGCGCGGCGGCUGGGUUGGGUUGGGUUG ......(((.(((((..(((..((.(((......)))))....(((((((.----((.(((.......))).)).)))))...((((...)))).)))))...)))))....))).. ( -36.40, z-score = 1.05, R) >consensus CUAAUCGCCAGCAGCAUGCCAGUGACCCUUUUAUGGUCACCAUGCAACGGGCACCGAUC_____CAAU__G_UC__CGAUCCGAAGCGACCCAAGUCGUCUUGAUUGGAUCGGCUUG ......(((....(((((...((((((.......)))))))))))................................(((((((((((((....)))).))...))))))))))... (-17.61 = -19.96 + 2.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:47 2011