| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,884,613 – 14,884,717 |
| Length | 104 |
| Max. P | 0.947528 |

| Location | 14,884,613 – 14,884,717 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.48793 |
| G+C content | 0.48043 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -15.29 |
| Energy contribution | -18.72 |
| Covariance contribution | 3.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
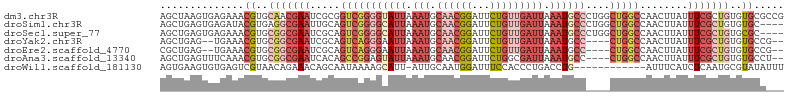
>dm3.chr3R 14884613 104 - 27905053 AGCUAAGUGAGAAACGUGCAACGAAUCGCGGUCGGGGUAUUAAAUGCAACGGAUUCUGUUGAUUAAAUGCCCUGGCUGGCCAACUUAUUUCGCUGUGUGCGCCG .((..(((((((..((.....)).....(((((((((((((.(((.((((((...))))))))).))))))))))))).........)))))))....)).... ( -36.50, z-score = -2.24, R) >droSim1.chr3R 20925182 100 + 27517382 AGCUGAGUGAGAUACGUGAGGCGAAUUGCAGUCGGGGCAUUAAAUGCAACGGAUUCUGUUGAUUAAAUGCCCUGGCUGGCCAACUUAUUUCGCUGUGUGC---- .((..(((((((((.((..(((......(((((((((((((.(((.((((((...))))))))).)))))))))))))))).)).)))))))))..))..---- ( -43.50, z-score = -5.02, R) >droSec1.super_77 63241 100 + 117255 AGCUGAGUGAGAAACGUGCGGCGAAUCGCAGUCGGGGCAUUAAAUGCAACGGAUUCUGUUGAUUAAAUGCCCUGGCUGGCCAACUUAUUUCGCUGUGCGC---- ..............((..(((((((...(((((((((((((.(((.((((((...))))))))).)))))))))))))..........)))))))..)).---- ( -42.72, z-score = -3.82, R) >droYak2.chr3R 3565467 96 - 28832112 AGCUGAG--UGAAACGUGCGGCGAAUCGCAGUCAGGGAAUUAAAUGCAACGGAUUCUGUUGAUUAAAUGCC----CUGGCCAACUUAUUUCGCUGUGUGCCG-- .......--.....((..(((((((.....(((((((.(((.(((.((((((...))))))))).))).))----)))))........)))))))..))...-- ( -31.72, z-score = -1.92, R) >droEre2.scaffold_4770 11001878 96 + 17746568 CGCUGAG--UGAAACGUGCGGCGAAUCGCAGUCAGGGAAUUAAAUGCAACGGAUUCUGUUGAUUAAAUGCC----CUGGCCAACUUAUUUCGCUGUGUGCCG-- .......--.....((..(((((((.....(((((((.(((.(((.((((((...))))))))).))).))----)))))........)))))))..))...-- ( -31.72, z-score = -1.88, R) >droAna3.scaffold_13340 17520854 98 - 23697760 AGCUGAGUUUCAAACGUGCGGCGAAUCACAGCCGGAGUAUUAAAUGCAACGGAUUCUGGCGAUUAAAUGCC----CUGGCCAACUUAUUUCGCUGUGUGCCU-- ..............((..(((((((.....(((((.(((((.(((((..(((...))))).))).))))).----)))))........)))))))..))...-- ( -26.52, z-score = 0.04, R) >droWil1.scaffold_181130 14565541 91 + 16660200 AGUGAAGUGUGAGUCGUAACAGAAACAGCAAUAAAAGCAUU-AUUGCAAUGGAUUUCCACCCUGACCUG------------AUUUCAUCUCAAUGCGUAUAUUU ..(((..((.((((((...(((.....(((((((.....))-)))))..(((....)))..)))...))------------))))))..)))............ ( -18.50, z-score = -1.04, R) >consensus AGCUGAGUGUGAAACGUGCGGCGAAUCGCAGUCGGGGCAUUAAAUGCAACGGAUUCUGUUGAUUAAAUGCC____CUGGCCAACUUAUUUCGCUGUGUGCC___ ..............((..(((((((.....(((((((((((.(((.((((((...))))))))).))))))....)))))........)))))))..))..... (-15.29 = -18.72 + 3.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:44 2011