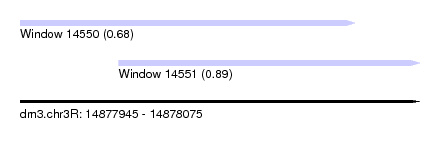
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,877,945 – 14,878,075 |
| Length | 130 |
| Max. P | 0.886563 |

| Location | 14,877,945 – 14,878,054 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.80 |
| Shannon entropy | 0.48214 |
| G+C content | 0.43850 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -16.09 |
| Energy contribution | -15.14 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
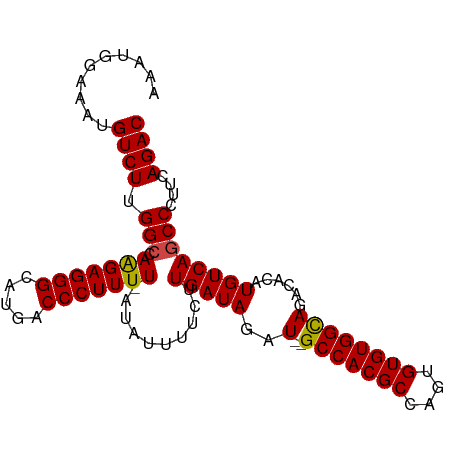
>dm3.chr3R 14877945 109 + 27905053 --ACCAUACACAAUGCGCUAUUACGCUGCAUAAAAAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUU-AUAUUUUCUUGAUAGAUAUGCCACGCCAGUGUGUGGCAG --.((((((((.((((((......)).))))............((.(.((((.(((((((.....)))))))-((((((.......)))))))))).).))))))))))... ( -32.00, z-score = -2.03, R) >droSim1.chr3R 20918721 101 - 27517382 ---------ACAAUGCGCUAUUACGCUGCAUAAAAAAUGAAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUUUAUAUUUUCUUGAUAGA--UGCCACGCCAGUGUGUGGCAG ---------...((((((......)).))))((((.(((((((.(((.((.((....)).)).)))..))))))).)))).........--((((((((....)))))))). ( -28.70, z-score = -1.75, R) >droSec1.super_77 56659 101 - 117255 ---------ACAAUGCGUUAUUACGCUGCAUAAAAAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUUUAUAUUUUCUUGAUAGA--UGCCACGCCAGUGUGUGGUAG ---------.....((((....))))..((..(((((((((((.(((.((.((....)).)).)))..))))))).))))..)).....--((((((((....)))))))). ( -26.80, z-score = -1.20, R) >droYak2.chr3R 3556748 106 + 28832112 --GCCGUACACAAAGCGUUAUUACGCUGCAUAAAAAAUGGAAAUGUCUGGGCCAAGAGGGCAUGACCCUUUU--AAAUUUCUUGAUAGA--UGCCACGCUAGUGUGUGGCAG --(((((((((..(((((....)))))((((.......((((((((....)).(((((((.....)))))))--..))))))......)--))).......))))))))).. ( -33.82, z-score = -2.30, R) >droEre2.scaffold_4770 10995344 105 - 17746568 --ACCGUACACAAAG-GCUAUUACGCUGCAUAAAAAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUU--AAGUUUCUUGAUAGA--UGCCACGCCAGUGUGUGGCAG --........(((.(-((((..(((.(.(((.....))).)..)))..)))))(((((((.....)))))))--.......))).....--((((((((....)))))))). ( -29.90, z-score = -1.12, R) >droAna3.scaffold_13340 17514256 108 + 23697760 UCUGUGUCCACAAAGCGCUAUUACGUUGCAUAAAAAAUGAAGAUGUCUUGGACAGGAGGGCAUGACCCUUUU--GAGAUGCUUGAUAGA--UGCCACGCCAGUGUGUGGCAG ((((((....).((((((((..(((((.(((.....)))..)))))..))).((((((((.....)))))))--)...))))).)))))--((((((((....)))))))). ( -33.00, z-score = -1.55, R) >dp4.chr2 6809723 106 + 30794189 --AUUAUUCAAAGAGAGUUAUUACACUGCAUAAAGAAAAGAGAGAUUGAAAUGUGGAGGGCAUGACCCUUUC--GAUGGGAGUGUCUGU--CGGUGUGUGUGUGUGUGGCAG --..............(((((.((((.(((((....(..((.((((.......(((((((......))))))--)........)))).)--)..).)))))))))))))).. ( -24.46, z-score = -0.54, R) >droPer1.super_0 647162 105 + 11822988 --AUUAUUCAAAGAGAGUUAUUACACUGCAUAAA-AAAAAAGAGAUUGAAAUGUGGAGGGCAUGACCCUUUC--GAUGGGAGUGUCUGU--CGGUGUGUCUGUGUGUGGCAG --..............(((((.((((.(((((..-....................(((((.....)))))((--((((((....)))))--))))))))..))))))))).. ( -23.30, z-score = -0.26, R) >consensus __ACCAUACACAAAGCGCUAUUACGCUGCAUAAAAAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUU__GAUUUUCUUGAUAGA__UGCCACGCCAGUGUGUGGCAG ................(((((.(((((((...............((....)).(((((((.....))))))).........................)).)))))))))).. (-16.09 = -15.14 + -0.95)

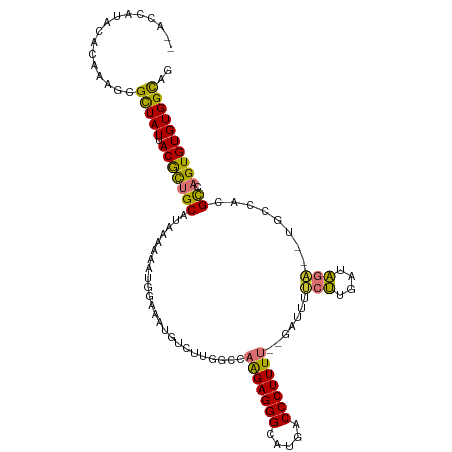
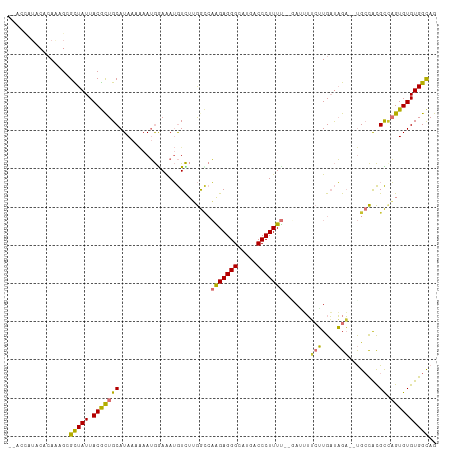
| Location | 14,877,977 – 14,878,075 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Shannon entropy | 0.12025 |
| G+C content | 0.47159 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.23 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14877977 98 + 27905053 AAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUU-AUAUUUUCUUGAUAGAUAUGCCACGCCAGUGUGUGGCAGACACAUGUCAGCCCUUCAGAC ..........((((.(((.(((((((.....)))))))-.........(((((....((((((((....))))))))......))))))))....)))) ( -30.20, z-score = -1.93, R) >droSim1.chr3R 20918746 97 - 27517382 AAAUGAAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUUUAUAUUUUCUUGAUAGAU--GCCACGCCAGUGUGUGGCAGACACAUGUCAGCCCUUCAGAC ..........((((.(((.(((((((.....)))))))..........(((((..(--(((((((....))))))))......))))))))....)))) ( -28.80, z-score = -1.93, R) >droSec1.super_77 56684 97 - 117255 AAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUUUAUAUUUUCUUGAUAGAU--GCCACGCCAGUGUGUGGUAGACACAUGUCAGCCCUUCAGAC ...(((..(.....)..))).((((((.((((.............(((....)))(--(((((((....)))))))).......))))))))))..... ( -27.10, z-score = -1.18, R) >droYak2.chr3R 3556780 95 + 28832112 AAAUGGAAAUGUCUGGGCCAAGAGGGCAUGACCCUUUU-A-AAUUUCUUGAUAGAU--GCCACGCUAGUGUGUGGCAGACACAUGUCAGCCCUUCAGAC ..........((((((((.(((((((.....)))))))-.-.......(((((..(--(((((((....))))))))......)))))))))...)))) ( -34.20, z-score = -3.00, R) >droEre2.scaffold_4770 10995375 95 - 17746568 AAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUU-A-AGUUUCUUGAUAGAU--GCCACGCCAGUGUGUGGCAGACACAUGUCAGCCCUUCAGAC ...(((..(.....)..))).((((((.((((.((.((-(-((...))))).)).(--(((((((....)))))))).......))))))))))..... ( -30.70, z-score = -1.90, R) >droAna3.scaffold_13340 17514290 95 + 23697760 AAAUGAAGAUGUCUUGGACAGGAGGGCAUGACCCUUUU--GAGAUGCUUGAUAGAU--GCCACGCCAGUGUGUGGCAGACACAUGUCAGCCCUUCAGAC ...(((((..((((....((((((((.....)))))))--)))))(((.((((..(--(((((((....))))))))......))))))).)))))... ( -33.90, z-score = -2.63, R) >consensus AAAUGGAAAUGUCUUGGCCAAGAGGGCAUGACCCUUUU_AUAUUUUCUUGAUAGAU__GCCACGCCAGUGUGUGGCAGACACAUGUCAGCCCUUCAGAC ...((((....((((....))))((((.((((.............(((....)))...(((((((....)))))))........))))))))))))... (-26.40 = -26.23 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:40 2011