
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,873,046 – 14,873,154 |
| Length | 108 |
| Max. P | 0.863666 |

| Location | 14,873,046 – 14,873,154 |
|---|---|
| Length | 108 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 61.52 |
| Shannon entropy | 0.78819 |
| G+C content | 0.56910 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -9.27 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14873046 108 + 27905053 --------GGACGUAUCGUUGAGCAAGUGGCUCUGCUGUU-GUUGCUGGGC-CGCAGCAGCG---GCUGCAGUUACGGCGGCAUUCGAUGUCGAGGUGGCGUAGUAGCAGCAGCGACACAA --------....((.((...((((.....)))).((((((-(((((((.((-(((....((.---((((......)))).)).((((....))))))))).))))))))))))))).)).. ( -51.90, z-score = -2.88, R) >droAna3.scaffold_13340 17509295 108 + 23697760 --------AGAGGUGUCGUUAAGAAGCUGGUUUUGGUGUU-GCUGCUGAGC-AGCAGCAGCU---GCCGCAGUUACUGCUGCAUUGGAUGUUGAGGUAGCGUAGUAACAGCAUCGGCACAG --------...(((((.((((....((((.(((.((((((-((((((....-))))))))).---)))(((((....)))))..........))).))))....)))).)))))....... ( -42.20, z-score = -1.28, R) >droEre2.scaffold_4770 10990375 108 - 17746568 --------GGACGUGUCGUUGAGCAAGUGGCUCGGCUGUU-GUUGUUGGAC-CGCAGCAGCG---GCUGCAGUUACGGCGGCGUUCGAUGUCGAGGUGGCAUAGUAGCAGCAGCGACACAA --------....(((((((((.((..((((((((((((((-(((((.....-.)))))))))---)))).)))))).((.((.((((....)))))).))......))..))))))))).. ( -52.70, z-score = -3.64, R) >droYak2.chr3R 3551067 108 + 28832112 --------GGACGUGUCGUUGAGCAAGUGGCUCUGCUGUU-GUUGUUGGAC-UGCAGCAGCG---GCUGCAGUUACGGCGGCGUUCGAUGUCGAGGUGGCAUAGUAGCAGCAGCGACACAA --------....(((((...((((.....)))).((((((-((((((((((-((((((....---)))))))))...((.((.((((....)))))).)).)))))))))))))))))).. ( -52.00, z-score = -3.35, R) >droSec1.super_77 51716 108 - 117255 --------GGACGUAUCGUUGAGCAAGUGGCUCUGCUGUU-GUUGCUGGGC-CGCUGCAGCG---GUUGCAGUUACUGCGGCAUUCGAUGUCGAGGUGGCGUAGUAGCAGCAGCGACACAA --------....((.((...((((.....)))).((((((-(((((((.((-((((((.(((---((.......))))).))..(((....))))))))).))))))))))))))).)).. ( -51.50, z-score = -2.91, R) >droSim1.chr3R 20913771 108 - 27517382 --------GGACGUAUCGUUGAGCAAGUGGCUCUGCUGUU-GUUGCUGGGC-CGCAGCAGCG---GCUGCAGUUACGGCGGCAUUCGAUGUCGAGGUGGCGUAGUAGCAGCAGCGACACAA --------....((.((...((((.....)))).((((((-(((((((.((-(((....((.---((((......)))).)).((((....))))))))).))))))))))))))).)).. ( -51.90, z-score = -2.88, R) >dp4.chr2 6804343 99 + 30794189 --------------------GGAGGUGUCAUUGAGCUGCU-GCUGCUGUUG-GGCAGCGGCGGCUGUAGCGGCCACAGCAGCAUUCGAUGUGGAAGUCGCAUAGUAGCAGCAUCGACACAG --------------------....(((((...((((((((-((((((((((-(((.((.((....)).)).))).))))))).....(((((.....)))))))))))))).))))))).. ( -46.50, z-score = -2.82, R) >droPer1.super_0 641872 99 + 11822988 --------------------GGAGGUGUCAUUGAGCUGCU-GCUGCUGUUG-GGCAGCGGCGGCUGUAGCGGCCACAGCAGCAUUCGAUGUGGAAGUCGCAUAGUAGCAGCAUCGACACAG --------------------....(((((...((((((((-((((((((((-(((.((.((....)).)).))).))))))).....(((((.....)))))))))))))).))))))).. ( -46.50, z-score = -2.82, R) >droWil1.scaffold_181130 14555401 93 - 16660200 --------------------GGAGGUGUCAUUAAGACGUU-GUUGCUGCUG-GGAAGC------UGCCGCAGUGACUGCAGCAUUUGAAGUGGAAGUUGCAUAAUAGCAGCAACGACACAA --------------------......(((.....)))(((-((((((((((-((..((------((...))))..))(((((.(((.....))).)))))....))))))))))))).... ( -33.10, z-score = -1.87, R) >droVir3.scaffold_12822 17181 93 + 4096053 --------------------GGGAGAAUCACUCUGAUGAU-GUUGUG---C-GACUGCAGCA---GCAGCCACUAUAGCGGUGGUUGAAGUAGGGGUUGCAUAGUAACAGCAACGACAAAG --------------------.((((.....)))).....(-((((((---(-((((.(.((.---.((((((((.....))))))))..)).).))))))...((....)).))))))... ( -31.40, z-score = -2.25, R) >droMoj3.scaffold_6540 32673034 96 + 34148556 --------------------GGGCGAGUCGCUGUGAUGUU-GUUGUGGCGU-CACGGCAGCA---GCAGCCACCAUGGCACUAGUGGACGAAGGCGUUGCAUAGUAGCAGCAUCGACACAG --------------------......((((((((((((((-.....)))))-)))))).((.---((.(((.....)))((((...((((....))))...)))).)).))...))).... ( -35.50, z-score = -0.51, R) >droGri2.scaffold_15074 4021235 87 - 7742996 --------------------AGGCGAAUCAUUAUGAUGAU-GAUGAU-------CGGCAACG---GCAGCUGC---GGCGACAAUUGAACCCGGCGUUGCAUAAUAGCAGCAACGACACAA --------------------.(.(((.(((((((....))-))))))-------)).)..((---...(((((---.(((((.............)))))......)))))..))...... ( -24.52, z-score = -0.81, R) >anoGam1.chr2R 51893808 117 + 62725911 UCAAUGUCUGGUUGGACAUCAGUCCGGUCAGUCCGUCGGCAGGCGUUCCACUCGACGCAGUA---CCUGGUCCUCCUGCACCGGUCAACGUUGCCGCUCCCGCGUAGAUGCAUCUGCACA- .....(.(((..(((((....)))))..))).).(.(((((((((((......))))).(.(---((.(((.(....).)))))))....)))))))....(.(((((....))))).).- ( -38.00, z-score = -0.06, R) >consensus _________G______C_U_GGGCAAGUCGCUCUGCUGUU_GUUGCUGGGC_CGCAGCAGCG___GCUGCAGUUACGGCGGCAUUCGAUGUCGAGGUGGCAUAGUAGCAGCAGCGACACAA .........................................((((((.........((.((....)).)).......((.((.(((......))))).)).....)))))).......... ( -9.27 = -9.55 + 0.29)

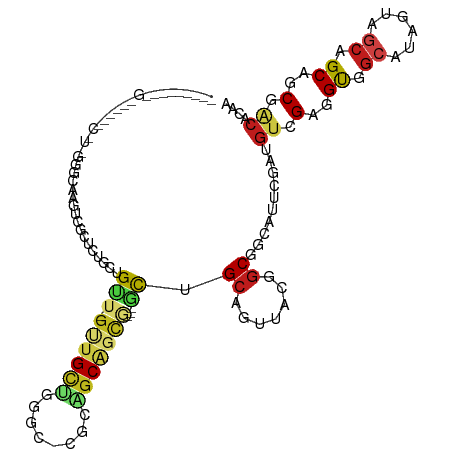
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:37 2011