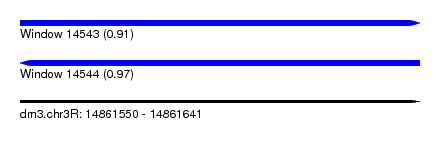
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,861,550 – 14,861,641 |
| Length | 91 |
| Max. P | 0.972438 |

| Location | 14,861,550 – 14,861,641 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.42279 |
| G+C content | 0.48770 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
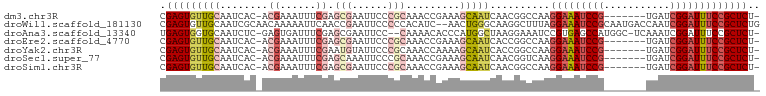
>dm3.chr3R 14861550 91 + 27905053 -AGAGCGGAAAUCCGAUCA-------CGGAUUUCCUUGGCCGUUGAUUGCUUUCGGUUUGCGGGAAUUCGCUCGAAAUUUCGU-GUGAUUGCAACACUCG -.(((((((((((((....-------)))))))))....((((.(((((....))))).))))......))))........((-((.......))))... ( -31.90, z-score = -2.04, R) >droWil1.scaffold_181130 14543751 98 - 16660200 CAGAGCGGAAAUCCGAUUGGUCAUUGCGGAUUUCCUAAAGCCUUGCCCAGUU--GAUGUGGGGGAAUUCGGUUGAAUUUUUGUUGCGAUUGCAACACUCG ..(((.(((((((((...........)))))))))...((((((.((((...--....)))).))....)))).......((((((....))))))))). ( -29.70, z-score = -0.97, R) >droAna3.scaffold_13340 17490288 95 + 23697760 -AGAGCGGAAAUCCGAUUUGA-GCCAUGGCUCACGGAUUUCCUUAGCCAUGGGUGUUUUG--GGAAUUCGCUCGAAAUCACUC-GAGAUUGCACCACUCA -.(((.(((((((((...(((-((....))))))))))))))...(((.(((((((((((--((......))))))).)))))-).)...))....))). ( -37.20, z-score = -3.58, R) >droEre2.scaffold_4770 10978775 91 - 17746568 -AGAGCGGAAAUCCGAUCA-------CGGAUUUCCUUGGCCGGUGAUUGCUUUCGGUUUGCGGGAAUUCGCUCGAAAUUUCGU-GUGAUUGCAACACUCG -.....(((((((((....-------))))))))).....(((((.((((..(((...((((..(.(((....))).)..)))-))))..))))))).)) ( -30.60, z-score = -1.38, R) >droYak2.chr3R 3539754 91 + 28832112 -AGAGCGGAAAUCCGAUCA-------CGGAUUUCCUUGGCCGGUGAUUGCUUUUGGUUUGCGGGAAUACAUUCGAAAUUUCGU-GUGAUUGCAACACUCG -.(((.(((((((((....-------))))))))).......(..((..(....(((((.((((......)))))))))....-)..))..)....))). ( -26.50, z-score = -0.97, R) >droSec1.super_77 40569 91 - 117255 -AGAGCGGAAAUCCGAUCA-------CGGAUUUCCUUGACCGUUGAUUGCUUUCGGUUUGCGGGAAUUUGCUCGAAAUUUCGU-GUGAUUGCAACACUCG -.(((((((((((((....-------)))))))))....((((.(((((....))))).))))......))))........((-((.......))))... ( -31.90, z-score = -2.70, R) >droSim1.chr3R 20904049 91 - 27517382 -AGAGCGGAAAUCCGAUCA-------CGGAUUUCCUUGGCCGUUGAUUGCUUUCGGUUUGCGGGAAUUCGCUCGAAAUUUCGU-GUGAUUGCAACACUCG -.(((((((((((((....-------)))))))))....((((.(((((....))))).))))......))))........((-((.......))))... ( -31.90, z-score = -2.04, R) >consensus _AGAGCGGAAAUCCGAUCA_______CGGAUUUCCUUGGCCGUUGAUUGCUUUCGGUUUGCGGGAAUUCGCUCGAAAUUUCGU_GUGAUUGCAACACUCG ..(((((((((((((...........)))))))))....((((.(((((....))))).))))......))))........................... (-17.80 = -18.77 + 0.97)

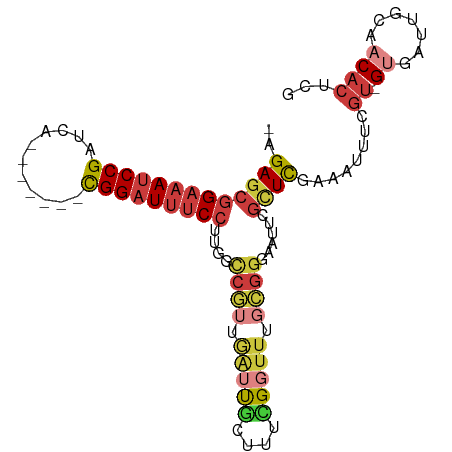
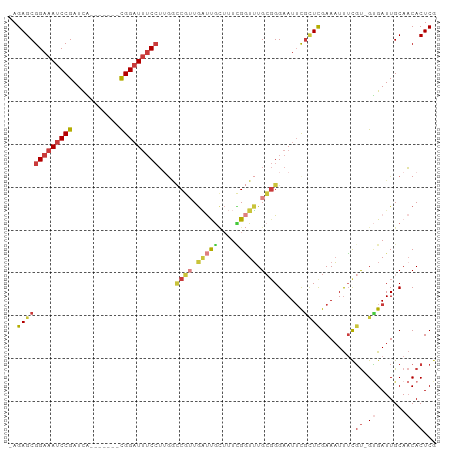
| Location | 14,861,550 – 14,861,641 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.42279 |
| G+C content | 0.48770 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -13.88 |
| Energy contribution | -15.50 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14861550 91 - 27905053 CGAGUGUUGCAAUCAC-ACGAAAUUUCGAGCGAAUUCCCGCAAACCGAAAGCAAUCAACGGCCAAGGAAAUCCG-------UGAUCGGAUUUCCGCUCU- .(((((((((..((..-.(((....))).(((......))).....))..)))))..........(((((((((-------....))))))))))))).- ( -29.20, z-score = -2.71, R) >droWil1.scaffold_181130 14543751 98 + 16660200 CGAGUGUUGCAAUCGCAACAAAAAUUCAACCGAAUUCCCCCACAUC--AACUGGGCAAGGCUUUAGGAAAUCCGCAAUGACCAAUCGGAUUUCCGCUCUG .(((((((((....)))))...(((((....)))))..((((....--...))))..........(((((((((...........))))))))))))).. ( -27.10, z-score = -2.43, R) >droAna3.scaffold_13340 17490288 95 - 23697760 UGAGUGGUGCAAUCUC-GAGUGAUUUCGAGCGAAUUCC--CAAAACACCCAUGGCUAAGGAAAUCCGUGAGCCAUGGC-UCAAAUCGGAUUUCCGCUCU- .((((((((....(((-(((....)))))).(......--)....)))).........((((((((((((((....))-)))...))))))))))))).- ( -35.00, z-score = -3.41, R) >droEre2.scaffold_4770 10978775 91 + 17746568 CGAGUGUUGCAAUCAC-ACGAAAUUUCGAGCGAAUUCCCGCAAACCGAAAGCAAUCACCGGCCAAGGAAAUCCG-------UGAUCGGAUUUCCGCUCU- ((.(((((((..((..-.(((....))).(((......))).....))..)))).))))).....(((((((((-------....))))))))).....- ( -29.50, z-score = -2.79, R) >droYak2.chr3R 3539754 91 - 28832112 CGAGUGUUGCAAUCAC-ACGAAAUUUCGAAUGUAUUCCCGCAAACCAAAAGCAAUCACCGGCCAAGGAAAUCCG-------UGAUCGGAUUUCCGCUCU- ((.(((((((......-.(((....)))..(((......)))........)))).))))).....(((((((((-------....))))))))).....- ( -25.30, z-score = -2.40, R) >droSec1.super_77 40569 91 + 117255 CGAGUGUUGCAAUCAC-ACGAAAUUUCGAGCAAAUUCCCGCAAACCGAAAGCAAUCAACGGUCAAGGAAAUCCG-------UGAUCGGAUUUCCGCUCU- ((.(((.......)))-.)).......((((......(((.....(....).......)))....(((((((((-------....))))))))))))).- ( -26.80, z-score = -2.48, R) >droSim1.chr3R 20904049 91 + 27517382 CGAGUGUUGCAAUCAC-ACGAAAUUUCGAGCGAAUUCCCGCAAACCGAAAGCAAUCAACGGCCAAGGAAAUCCG-------UGAUCGGAUUUCCGCUCU- .(((((((((..((..-.(((....))).(((......))).....))..)))))..........(((((((((-------....))))))))))))).- ( -29.20, z-score = -2.71, R) >consensus CGAGUGUUGCAAUCAC_ACGAAAUUUCGAGCGAAUUCCCGCAAACCGAAAGCAAUCAACGGCCAAGGAAAUCCG_______UGAUCGGAUUUCCGCUCU_ .(((((((((........((......)).(((......))).........)))))..........(((((((((...........))))))))))))).. (-13.88 = -15.50 + 1.62)
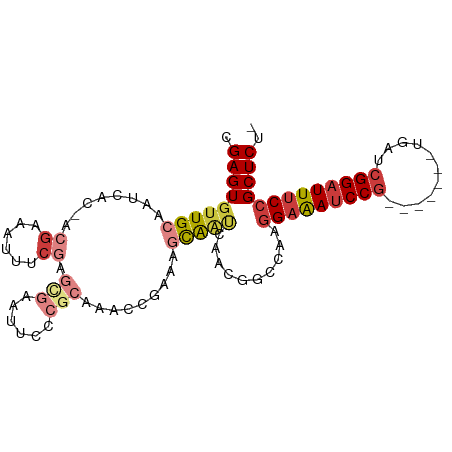
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:34 2011