
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,838,159 – 14,838,250 |
| Length | 91 |
| Max. P | 0.662080 |

| Location | 14,838,159 – 14,838,250 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 71.26 |
| Shannon entropy | 0.54936 |
| G+C content | 0.53222 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -11.47 |
| Energy contribution | -11.00 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14838159 91 - 27905053 UGUGGCUGCUCC-AUUUCGGAGUUU-----AAAAAGCCGAACAAAUGAG-CCAUGGCCGGGAUGGAGAUGGCGAUGGAAAUCGGUGGAGGGGUGUGUC ....(((.((((-(((((((..(..-----...)..)))))....(((.-((((.((((.........)))).))))...))))))))).)))..... ( -30.60, z-score = -2.11, R) >droSim1.chr3R_random 796717 92 - 1307089 UGUGGCUGCUCC-AUUUCGGAGUUA-----AAAAAGCCGAACAAAUGAGCCCAUGGCCGGGAUGGAGAAGGCGAUGGAAAUCGGUGGAGGGAAGUGUC ....(((.((((-(((((((..(..-----...)..)))))....(((..((((.(((...........))).))))...)))))))))...)))... ( -26.30, z-score = -0.71, R) >droSec1.super_77 17289 91 + 117255 UGUGGCUGCUCC-AUUUCGGAGUUU-----AAAAAGCCGAACAAAUGAG-CCAUGGCCGGGAUGGAGAUGGCGGUGGAAAUCGGUGGAGGGGAGUGUC .....((.((((-(((((((..(..-----...)..)))))....(((.-((((.((((.........)))).))))...))))))))).))...... ( -29.10, z-score = -1.57, R) >droYak2.chr3R 3516278 90 - 28832112 UGUGGCUGCUCC-AUUUCGGAGUUC-----AAAAAGCCGAACAAAUGAG-CCAUGGCCGGUAUGGAGAUGG-GAUGGAAAUCGAAGGAGGGGUGUGUC ....(((.((((-..(((((..(..-----...)..)))))........-((((..((.....))..))))-(((....)))...)))).)))..... ( -24.50, z-score = -0.37, R) >droEre2.scaffold_4770 10955275 91 + 17746568 UGUGGCUGCUCC-AUUUCGGAGUUU-----AAAAAGCCGAGCAAAUGAG-CCAUGGCCAGUAUGGAUAUGGAGAUGGAAAUCGUAGGAGGGGUGUGUU ....(((.((((-..(((((..(..-----...)..)))))...((((.-((((..(((.........)))..))))...)))).)))).)))..... ( -26.40, z-score = -1.42, R) >droAna3.scaffold_12911 3198543 75 - 5364042 UGUGGCUGCUCC-AUUUCGGAGUUU--UAAAAAACAUCGAACAAAUGAG-CCAUGGCCGCAUCGGACACGCCCAGUGUC------------------- (((((((((((.-..((((..((((--....))))..)))).....)))-)...)))))))...(((((.....)))))------------------- ( -25.50, z-score = -2.65, R) >droWil1.scaffold_181089 4130627 81 + 12369635 UGCCGCCGCUCCAAUUUUGGGGCCAGUUGUGGCGCAUCAAACAAAUGAG-CCAUGGCCGCCACAAAUUGGGCGUGGCUGGUC---------------- .((((((((((((((((((((((((....((((.(((.......))).)-)))))))).))).)))))))).))))).))).---------------- ( -39.70, z-score = -2.89, R) >consensus UGUGGCUGCUCC_AUUUCGGAGUUU_____AAAAAGCCGAACAAAUGAG_CCAUGGCCGGGAUGGAGAUGGCGAUGGAAAUCGGUGGAGGGGUGUGUC ..((((((((((......))))).....................(((....))))))))....................................... (-11.47 = -11.00 + -0.47)

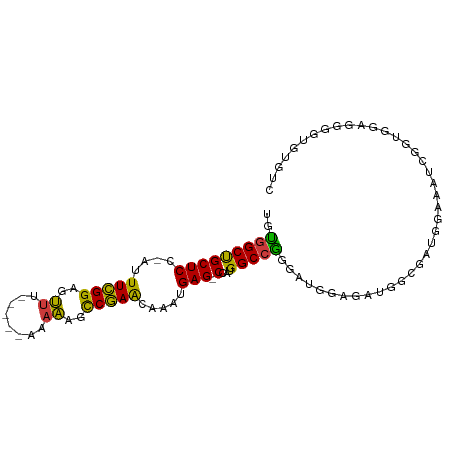
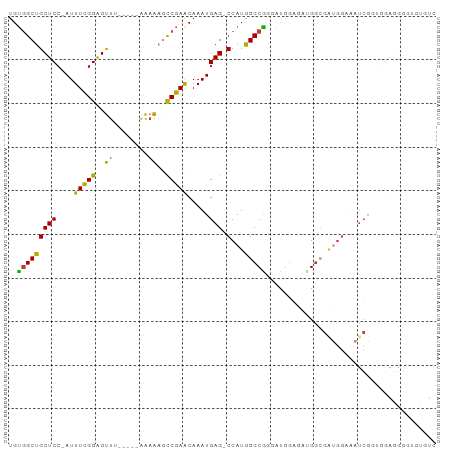
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:33 2011