| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,835,657 – 14,835,769 |
| Length | 112 |
| Max. P | 0.795452 |

| Location | 14,835,657 – 14,835,769 |
|---|---|
| Length | 112 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Shannon entropy | 0.49544 |
| G+C content | 0.47054 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -14.42 |
| Energy contribution | -15.36 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 14835657 112 + 27905053 UCUUGUUUAUCUUUUAGCCGGGCCAUUAGAUAAGCUGCGCAAAGACAAACAGCCAAUGGCCAAUAUCUGCAGACGGCGAUGAUGCCAAUGGCAACCAACAGUAACUCUGCAG ....((((((((..(.((...)).)..))))))))...(((.((.......((((.((((...((((.((.....))))))..)))).)))).((.....))..)).))).. ( -28.00, z-score = -0.17, R) >droSim1.chr3R 20893909 112 - 27517382 UCUUGUUUAUCUUUUAGCCGGGCCAUUAGAUAAGCUGUGCAAAGACAAACAGCCAAUGGCCAAUAUCUGCAGACGGCGAUGAUGCCAAUGGCAACCAACAGUAACUCUGCAA ....((((((((..(.((...)).)..))))))))..((((.((.......((((.((((...((((.((.....))))))..)))).)))).((.....))..)).)))). ( -28.60, z-score = -0.32, R) >droSec1.super_77 14822 112 - 117255 UCUUGUUUAUCUUUUAGCCGGGCCAUUAGAUAAGCUGUGCAAAGACAAACAGCCAAUGGCCAAUAUCUGCAGACGGCGAUGAUGCCAAUGGCAACCAACAGUAACUCUGCCA .......((((.....(((((((((((.(....(((((..........)))))))))))))....((....))))))...))))....(((((..............))))) ( -27.84, z-score = -0.02, R) >droYak2.chr3R 3513822 112 + 28832112 UCUUGUUUAUUUUUUAGCCGGGCCAUUAGAUAAGCUGUGCAAAGACAAACAGCCAAUGGCCAAUAUCUGCAGACGGCGAUGAUGCCAAUGGCAACGAACAGUAACUCUGCAA ..((((((........(((((((((((.(....(((((..........)))))))))))))....((....)))))).....((((...))))..))))))........... ( -29.10, z-score = -0.57, R) >droEre2.scaffold_4770 10952769 112 - 17746568 UCUUGUUUAUCUUUUAGCCGGGCCAUUAGAUAAGCUGUGCAAAGACAAACAGCCAAUGGCCAAUAUCUGCAGACGGCGAUGAUGCCAAUGGCAACGAACAGUAACUCUGCAA ....((((((((..(.((...)).)..))))))))..((((.((.......((((.((((...((((.((.....))))))..)))).)))).((.....))..)).)))). ( -29.80, z-score = -0.70, R) >droAna3.scaffold_12911 3196020 86 + 5364042 GCUUGUUUAUCUUUUAGCCAGGCCAUUAGAUAAGCUCCGCAAAGACAAACAGCCAGCGGCCAAUG---------GACUGGGAUGCCAAUGGCAAC----------------- ((((((((.(((((..((..(((..........)))..))))))).)))))...))).((((.((---------(.(......)))).))))...----------------- ( -22.40, z-score = 0.01, R) >dp4.chr2 14958919 84 + 30794189 UCCUGUUUAUCUGCUGGCCAGGCCAUUAGAUAAGCUCCA-AAAGACAAACAGCCAAUGGCCAAUA---------GAC---GAUGCCAUUGGCAAAGA--------------- ....(((((((((.((((...)))).)))))))))....-...........(((((((((.....---------...---...))))))))).....--------------- ( -30.20, z-score = -3.91, R) >droPer1.super_0 6371593 84 - 11822988 UCCAGUUUAUCUGCUGGCCAGGCCAUUAGAUAAGCUCCA-AAAGACAAACAGCCAAUGGCCAAUA---------GAC---GAUGCCAUUGGCAAAGA--------------- ...((((((((((.((((...)))).))))))))))...-...........(((((((((.....---------...---...))))))))).....--------------- ( -31.00, z-score = -4.51, R) >droMoj3.scaffold_6540 28860792 86 + 34148556 -------CCUUGUUUAUCCAUGCCAUUAGAUAAGCUG-GCCAAGACAAACAGCCAGUGGCCAAAGGCUGACGACGACAAGGUGGCGAAUGGGAA------------------ -------.........(((((((((((......((((-((...........))))))((((...))))...........))))))..)))))..------------------ ( -23.70, z-score = -0.57, R) >consensus UCUUGUUUAUCUUUUAGCCGGGCCAUUAGAUAAGCUGCGCAAAGACAAACAGCCAAUGGCCAAUAUCUGCAGACGGCGAUGAUGCCAAUGGCAACGAACAGUAACUCUGC__ ....((((((((..(.((...)).)..))))))))................((((.((((.......................)))).)))).................... (-14.42 = -15.36 + 0.94)

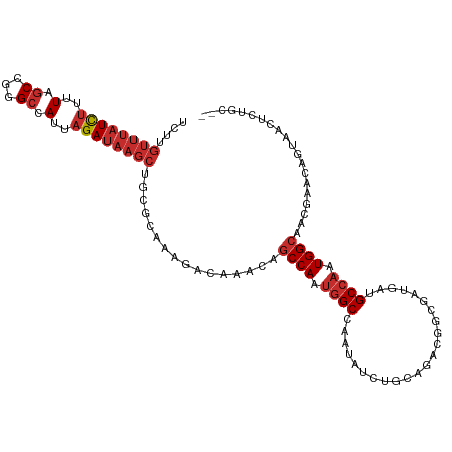
| Location | 14,835,657 – 14,835,769 |
|---|---|
| Length | 112 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Shannon entropy | 0.49544 |
| G+C content | 0.47054 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.19 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14835657 112 - 27905053 CUGCAGAGUUACUGUUGGUUGCCAUUGGCAUCAUCGCCGUCUGCAGAUAUUGGCCAUUGGCUGUUUGUCUUUGCGCAGCUUAUCUAAUGGCCCGGCUAAAAGAUAAACAAGA (((((((((.((.....)).))....(((......))).))))))).....((((...))))((((((((((..((.(((........)))...))..)))))))))).... ( -34.30, z-score = -0.62, R) >droSim1.chr3R 20893909 112 + 27517382 UUGCAGAGUUACUGUUGGUUGCCAUUGGCAUCAUCGCCGUCUGCAGAUAUUGGCCAUUGGCUGUUUGUCUUUGCACAGCUUAUCUAAUGGCCCGGCUAAAAGAUAAACAAGA ..((((.....)))).....((((.((((...(((((.....)).)))....)))).))))(((((((((((((...(((........)))...))..)))))))))))... ( -33.10, z-score = -0.62, R) >droSec1.super_77 14822 112 + 117255 UGGCAGAGUUACUGUUGGUUGCCAUUGGCAUCAUCGCCGUCUGCAGAUAUUGGCCAUUGGCUGUUUGUCUUUGCACAGCUUAUCUAAUGGCCCGGCUAAAAGAUAAACAAGA .(((((.....)))))....((((.((((...(((((.....)).)))....)))).))))(((((((((((((...(((........)))...))..)))))))))))... ( -34.20, z-score = -0.65, R) >droYak2.chr3R 3513822 112 - 28832112 UUGCAGAGUUACUGUUCGUUGCCAUUGGCAUCAUCGCCGUCUGCAGAUAUUGGCCAUUGGCUGUUUGUCUUUGCACAGCUUAUCUAAUGGCCCGGCUAAAAAAUAAACAAGA (((((((((.((.....)).))....(((......))).))))))).....(((((((((((((..........)))))).....))))))).................... ( -28.60, z-score = 0.10, R) >droEre2.scaffold_4770 10952769 112 + 17746568 UUGCAGAGUUACUGUUCGUUGCCAUUGGCAUCAUCGCCGUCUGCAGAUAUUGGCCAUUGGCUGUUUGUCUUUGCACAGCUUAUCUAAUGGCCCGGCUAAAAGAUAAACAAGA ..((((.....)))).....((((.((((...(((((.....)).)))....)))).))))(((((((((((((...(((........)))...))..)))))))))))... ( -33.10, z-score = -1.01, R) >droAna3.scaffold_12911 3196020 86 - 5364042 -----------------GUUGCCAUUGGCAUCCCAGUC---------CAUUGGCCGCUGGCUGUUUGUCUUUGCGGAGCUUAUCUAAUGGCCUGGCUAAAAGAUAAACAAGC -----------------((.((((.((((......).)---------)).)))).))..(((((((((((((.(((.(((........))))))....)))))))))).))) ( -28.60, z-score = -1.33, R) >dp4.chr2 14958919 84 - 30794189 ---------------UCUUUGCCAAUGGCAUC---GUC---------UAUUGGCCAUUGGCUGUUUGUCUUU-UGGAGCUUAUCUAAUGGCCUGGCCAGCAGAUAAACAGGA ---------------.....(((((((((...---...---------.....)))))))))(((((((((.(-(((.(((........)))....)))).)))))))))... ( -31.80, z-score = -2.82, R) >droPer1.super_0 6371593 84 + 11822988 ---------------UCUUUGCCAAUGGCAUC---GUC---------UAUUGGCCAUUGGCUGUUUGUCUUU-UGGAGCUUAUCUAAUGGCCUGGCCAGCAGAUAAACUGGA ---------------.....(((((((((...---...---------.....))))))))).((((((((.(-(((.(((........)))....)))).)))))))).... ( -30.50, z-score = -2.58, R) >droMoj3.scaffold_6540 28860792 86 - 34148556 ------------------UUCCCAUUCGCCACCUUGUCGUCGUCAGCCUUUGGCCACUGGCUGUUUGUCUUGGC-CAGCUUAUCUAAUGGCAUGGAUAAACAAGG------- ------------------.............((((((........((...((((((..(((.....))).))))-))))(((((((......)))))))))))))------- ( -24.10, z-score = -0.99, R) >consensus __GCAGAGUUACUGUUCGUUGCCAUUGGCAUCAUCGCCGUCUGCAGAUAUUGGCCAUUGGCUGUUUGUCUUUGCACAGCUUAUCUAAUGGCCCGGCUAAAAGAUAAACAAGA ....................(((...(((......))).............(((((((((..(((((......)).)))....))))))))).)))................ (-19.24 = -19.19 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:32 2011