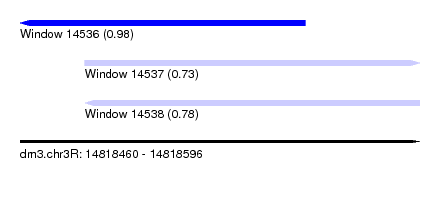
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,818,460 – 14,818,596 |
| Length | 136 |
| Max. P | 0.977310 |

| Location | 14,818,460 – 14,818,557 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Shannon entropy | 0.18171 |
| G+C content | 0.48308 |
| Mean single sequence MFE | -13.31 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
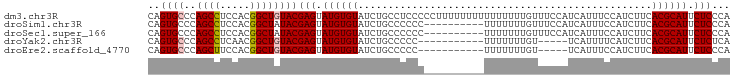
>dm3.chr3R 14818460 97 - 27905053 CAGUGCCCAGCCUCCACGGCUGUACGAGUAUGUGUAUCUGCCUCCCCCUUUUUUUUUUUUUUUGUUUCCAUCAUUUCCAUCUUCACGCAUUCUCCCA ..((((..((((.....))))))))(((.((((((.................................................)))))).)))... ( -14.79, z-score = -2.97, R) >droSim1.chr3R 20864239 87 + 27517382 CAGUGCCCAGCCUCCACGGCUAUACGAGUAUGUGUAUCUGCCCCCC----------UUUUUUUGUUUCCAUCAUUUCCAUCUUCACGCAUUCUCCCA ..(((...((((.....)))).)))(((.((((((...........----------............................)))))).)))... ( -11.73, z-score = -1.53, R) >droSec1.super_166 33072 87 + 36471 CAGUGCCCAGCCUCCACGGCUAUACGAGUAUGUGUAUCUGCCCCCC----------UUUUUUUGUUUCCAUCAUUUCCAUCUUCACGCAUUCUCCCA ..(((...((((.....)))).)))(((.((((((...........----------............................)))))).)))... ( -11.73, z-score = -1.53, R) >droYak2.chr3R 3502078 81 - 28832112 CAGUGCCCAGCCUCAACGGCUGUACGAGUAUGUGUAUCUGCCCCC-----------UUUUUUUGU-----UCAUUUUCAUCUUCACGCAUUCUCUCA ..((((..((((.....))))))))(((.((((((..........-----------.........-----..............)))))).)))... ( -15.61, z-score = -2.23, R) >droEre2.scaffold_4770 10935523 81 + 17746568 CAGUGCCCAGCUUCCACGGCUGUACGAGUAUGUGUAUCUGCCCCC-----------UUUUUUUGU-----UCAUUUCCAUCUUCACGCAUUCUCCCA ..((((..((((.....))))))))(((.((((((..........-----------.........-----..............)))))).)))... ( -12.71, z-score = -1.28, R) >consensus CAGUGCCCAGCCUCCACGGCUGUACGAGUAUGUGUAUCUGCCCCCC__________UUUUUUUGUUUCCAUCAUUUCCAUCUUCACGCAUUCUCCCA ..((((..((((.....))))))))(((.((((((.................................................)))))).)))... (-12.93 = -13.17 + 0.24)
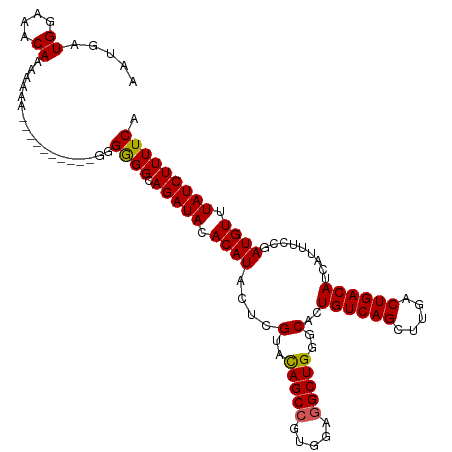
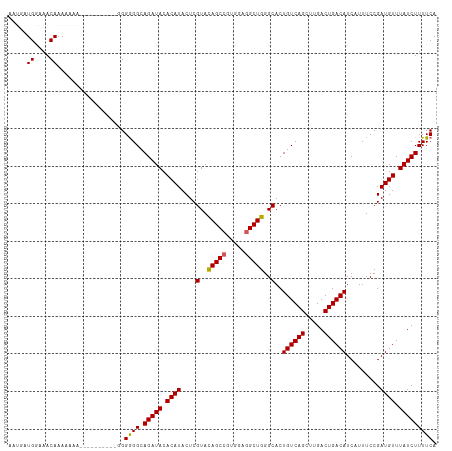
| Location | 14,818,482 – 14,818,596 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Shannon entropy | 0.15046 |
| G+C content | 0.43664 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.74 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
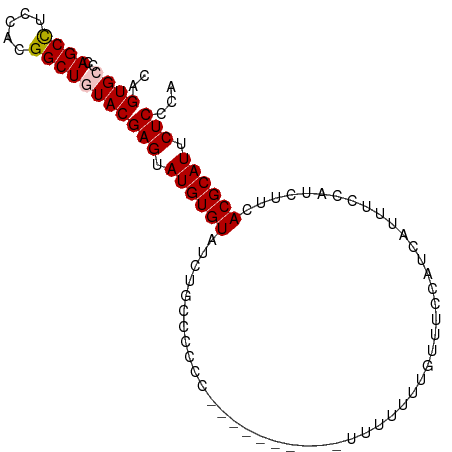
>dm3.chr3R 14818482 114 + 27905053 AAUGAUGGAAACAAAAAAAAAAAAAAAGGGGGAGGCAGAUACACAUACUCGUACAGCCGUGGAGGCUGGGCACUGUCAGCUUGACUGACAUCAUUUCCGAUGUUUAUCUUUUCA .....((....))..................((((.(((((.((((....(..(((((.....)))))..)..((((((.....)))))).........)))).))))))))). ( -29.80, z-score = -1.72, R) >droSim1.chr3R 20864261 104 - 27517382 AAUGAUGGAAACAAAAAAA----------GGGGGGCAGAUACACAUACUCGUAUAGCCGUGGAGGCUGGGCACUGUCAGCUUGACUGACAUCAUUUCCGAUGUUUAUCUUUUCA .....((....))......----------..(..(.(((((.((((....((.(((((.....))))).))..((((((.....)))))).........)))).))))))..). ( -27.60, z-score = -1.19, R) >droSec1.super_166 33094 104 - 36471 AAUGAUGGAAACAAAAAAA----------GGGGGGCAGAUACACAUACUCGUAUAGCCGUGGAGGCUGGGCACUGUCAGCUUGACUGACAUCAUUUCCGAUGUUUAUCUUUUCA .....((....))......----------..(..(.(((((.((((....((.(((((.....))))).))..((((((.....)))))).........)))).))))))..). ( -27.60, z-score = -1.19, R) >droYak2.chr3R 3502100 98 + 28832112 ---AAUG--AACAAAAAAA-----------GGGGGCAGAUACACAUACUCGUACAGCCGUUGAGGCUGGGCACUGUCAGCUUGACUGACAUCAUUUCCGAUGUUUAUCUUUUCA ---....--..........-----------.(..(.(((((.((((....(..(((((.....)))))..)..((((((.....)))))).........)))).))))))..). ( -27.30, z-score = -1.89, R) >droEre2.scaffold_4770 10935545 98 - 17746568 ---AAUG--AACAAAAAAA-----------GGGGGCAGAUACACAUACUCGUACAGCCGUGGAAGCUGGGCACUGUCAGCUUGACUGACAUCAUUUCCGAUGUUUAUCUUUUCA ---....--..........-----------.(..(.(((((.((((....(..((((.......))))..)..((((((.....)))))).........)))).))))))..). ( -22.50, z-score = -0.34, R) >consensus AAUGAUGGAAACAAAAAAA__________GGGGGGCAGAUACACAUACUCGUACAGCCGUGGAGGCUGGGCACUGUCAGCUUGACUGACAUCAUUUCCGAUGUUUAUCUUUUCA .....((....))..................((((.(((((.((((....(..(((((.....)))))..)..((((((.....)))))).........)))).))))))))). (-24.94 = -24.74 + -0.20)

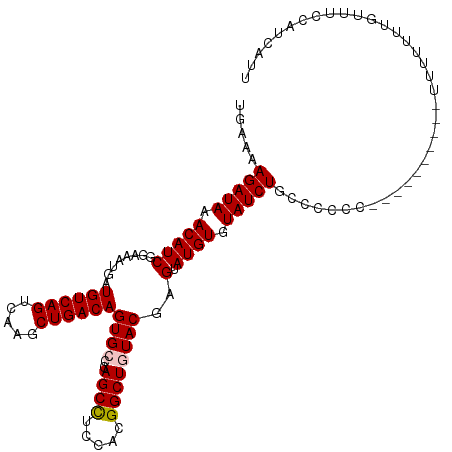
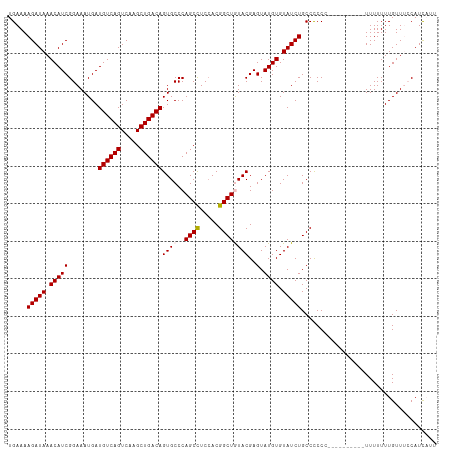
| Location | 14,818,482 – 14,818,596 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.69 |
| Shannon entropy | 0.15046 |
| G+C content | 0.43664 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14818482 114 - 27905053 UGAAAAGAUAAACAUCGGAAAUGAUGUCAGUCAAGCUGACAGUGCCCAGCCUCCACGGCUGUACGAGUAUGUGUAUCUGCCUCCCCCUUUUUUUUUUUUUUUGUUUCCAUCAUU (((...(((....)))(((((..(((((((.....))))))((((..((((.....))))))))....................................)..))))).))).. ( -25.40, z-score = -1.57, R) >droSim1.chr3R 20864261 104 + 27517382 UGAAAAGAUAAACAUCGGAAAUGAUGUCAGUCAAGCUGACAGUGCCCAGCCUCCACGGCUAUACGAGUAUGUGUAUCUGCCCCCC----------UUUUUUUGUUUCCAUCAUU (((...(((....)))(((((..(((((((.....))))))(((.........)))(((.(((((......)))))..)))....----------.....)..))))).))).. ( -23.20, z-score = -1.44, R) >droSec1.super_166 33094 104 + 36471 UGAAAAGAUAAACAUCGGAAAUGAUGUCAGUCAAGCUGACAGUGCCCAGCCUCCACGGCUAUACGAGUAUGUGUAUCUGCCCCCC----------UUUUUUUGUUUCCAUCAUU (((...(((....)))(((((..(((((((.....))))))(((.........)))(((.(((((......)))))..)))....----------.....)..))))).))).. ( -23.20, z-score = -1.44, R) >droYak2.chr3R 3502100 98 - 28832112 UGAAAAGAUAAACAUCGGAAAUGAUGUCAGUCAAGCUGACAGUGCCCAGCCUCAACGGCUGUACGAGUAUGUGUAUCUGCCCCC-----------UUUUUUUGUU--CAUU--- .....(((((.(((((((......((((((.....))))))...))(((((.....))))).....).)))).)))))......-----------..........--....--- ( -24.10, z-score = -1.49, R) >droEre2.scaffold_4770 10935545 98 + 17746568 UGAAAAGAUAAACAUCGGAAAUGAUGUCAGUCAAGCUGACAGUGCCCAGCUUCCACGGCUGUACGAGUAUGUGUAUCUGCCCCC-----------UUUUUUUGUU--CAUU--- ((((..(((..((((((....))))))..)))((((((........))))))....(((.(((((......)))))..)))...-----------........))--))..--- ( -22.20, z-score = -0.77, R) >consensus UGAAAAGAUAAACAUCGGAAAUGAUGUCAGUCAAGCUGACAGUGCCCAGCCUCCACGGCUGUACGAGUAUGUGUAUCUGCCCCCC__________UUUUUUUGUUUCCAUCAUU .....(((((.(((((........((((((.....))))))((((..((((.....))))))))..).)))).))))).................................... (-22.06 = -22.30 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:29 2011