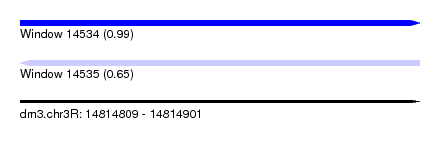
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,814,809 – 14,814,901 |
| Length | 92 |
| Max. P | 0.986771 |

| Location | 14,814,809 – 14,814,901 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.63205 |
| G+C content | 0.53776 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.05 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
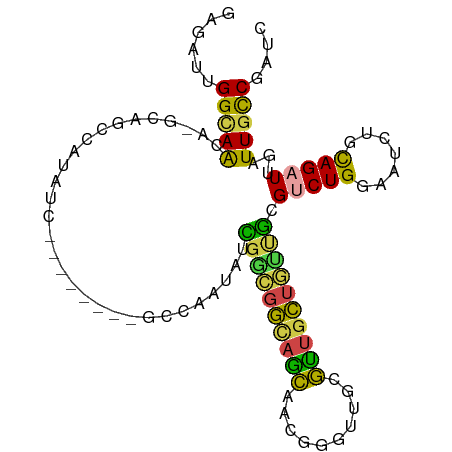
>dm3.chr3R 14814809 92 + 27905053 GAGAUUGGCAAGA-GCAGCCAUAUC--------GCCAAUAUCGACGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUC ..(((((((((..-(((((......--------(((.........)))(((((((.....))))))))))))(((((.......)))))...))))))))) ( -35.40, z-score = -2.10, R) >droSim1.chr3R 20860206 100 - 27517382 GAGAUUGGCAACA-GCAGCCAUAUCCCAAUAUCGCCAAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUC ..(((((((((((-(((...............((((......))))(((((((((.....))))))))))))(((((.......))))))).))))))))) ( -39.30, z-score = -2.43, R) >droSec1.super_166 29502 100 - 36471 GAGAUUGGCAACA-GCAGCCAUAUCCCAAUAUCGCCAAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUC ..(((((((((((-(((...............((((......))))(((((((((.....))))))))))))(((((.......))))))).))))))))) ( -39.30, z-score = -2.43, R) >droEre2.scaffold_4770 10931934 81 - 17746568 --GAAUGGCAACA-GCAGC-----------------AAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGUAGAUUGAUUGCCGAUC --((.((((((((-.....-----------------.......((((((((((((.....))))))))))))(((((.......))))))).)))))).)) ( -29.90, z-score = -1.78, R) >dp4.chr2 14934669 83 + 30794189 GAGACGGGGAGGA-GAGGGAGAG----------GAGAAGAUUGCUUGUUGC-----GUGCCGCUCCU--CGCGACUGGAAUCUGCAGAUUGAUUGCCGAUC ......(.(((((-(.((.((..----------((.(((....))).)).)-----.).)).)))))--).)((.(((((((........)))).))).)) ( -22.30, z-score = 0.81, R) >droWil1.scaffold_181089 4103082 101 - 12369635 GGAAGUGGUAAGAUGCAGUUUGGUCUGGGUCUGGCUAUGGCUAUGGCUAUGGGCCUGGGCCUAUGGUUGGACGACUGGAAUCUGCAGAUUGAUUGUCGAUC .((.....(((..(((((((..(((...(((..(((((((((..(((.....)))..)))).)))))..))))))..))..)))))..)))....)).... ( -39.10, z-score = -3.51, R) >consensus GAGAUUGGCAACA_GCAGCCAUAUC________GCCAAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUC ......(((((..............................((((((((((..........)))))))))).(((((.......)))))...))))).... (-16.12 = -15.05 + -1.07)

| Location | 14,814,809 – 14,814,901 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.63205 |
| G+C content | 0.53776 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -8.06 |
| Energy contribution | -11.17 |
| Covariance contribution | 3.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
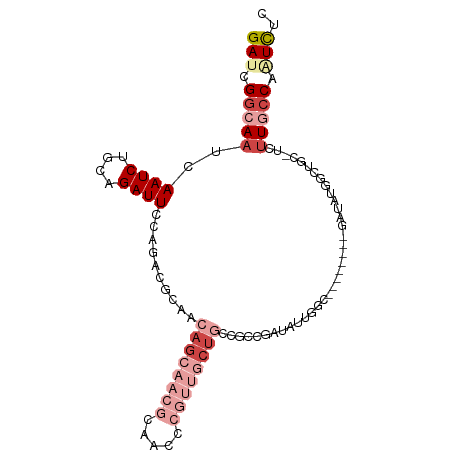
>dm3.chr3R 14814809 92 - 27905053 GAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGUCGAUAUUGGC--------GAUAUGGCUGC-UCUUGCCAAUCUC .((((.((((.((((....))))...((((...((((((((.....)))))))).))))...)))).)--------))).((((...-....))))..... ( -30.30, z-score = -2.10, R) >droSim1.chr3R 20860206 100 + 27517382 GAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUGGGAUAUGGCUGC-UGUUGCCAAUCUC (((.((((((......((..(((((((......((((((((.....)))))))).(((((....)))))...)))))))...))...-.)))))).))).. ( -37.50, z-score = -2.67, R) >droSec1.super_166 29502 100 + 36471 GAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUGGGAUAUGGCUGC-UGUUGCCAAUCUC (((.((((((......((..(((((((......((((((((.....)))))))).(((((....)))))...)))))))...))...-.)))))).))).. ( -37.50, z-score = -2.67, R) >droEre2.scaffold_4770 10931934 81 + 17746568 GAUCGGCAAUCAAUCUACAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUU-----------------GCUGC-UGUUGCCAUUC-- ....(((((..((((....)))).(((..((((((((((((.....)))))))).........))-----------------))..)-)))))))....-- ( -23.80, z-score = -2.12, R) >dp4.chr2 14934669 83 - 30794189 GAUCGGCAAUCAAUCUGCAGAUUCCAGUCGCG--AGGAGCGGCAC-----GCAACAAGCAAUCUUCUC----------CUCUCCCUC-UCCUCCCCGUCUC (((.((.((((........)))))).)))..(--(((((.((...-----((.....))...)).)))----------)))......-............. ( -18.70, z-score = -0.66, R) >droWil1.scaffold_181089 4103082 101 + 12369635 GAUCGACAAUCAAUCUGCAGAUUCCAGUCGUCCAACCAUAGGCCCAGGCCCAUAGCCAUAGCCAUAGCCAGACCCAGACCAAACUGCAUCUUACCACUUCC ((.((((....((((....))))...))))))........(((...(((.....)))...)))......(((..(((......)))..))).......... ( -17.10, z-score = -1.50, R) >consensus GAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGC________GAUAUGGCUGC_UCUUGCCAAUCUC (((.(((((..((((....))))..........((((((((.....))))))))....................................))))).))).. ( -8.06 = -11.17 + 3.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:27 2011