
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,796,639 – 14,796,713 |
| Length | 74 |
| Max. P | 0.999967 |

| Location | 14,796,639 – 14,796,713 |
|---|---|
| Length | 74 |
| Sequences | 6 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.60810 |
| G+C content | 0.45336 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -12.31 |
| Energy contribution | -12.76 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.58 |
| Mean z-score | -5.45 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.36 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
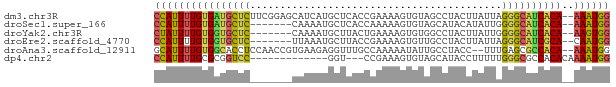
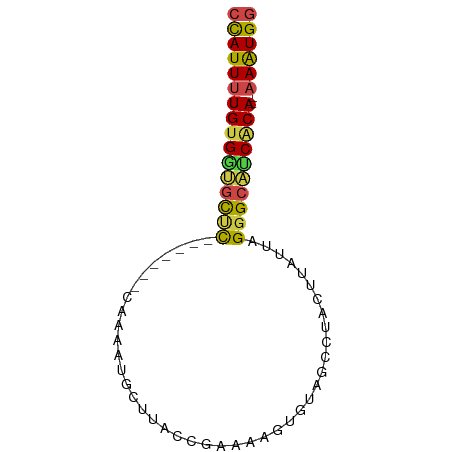
>dm3.chr3R 14796639 74 + 27905053 CCAUUUUGUGAUGCUCUUCGGAGCAUCAUGCUCACCGAAAAGUGUAGCCUACUUAUUAGGGCAUCACA--AAAUGG ((((((((((((((((((((((((.....)))..)))))(((((.....)))))....))))))))))--)))))) ( -34.20, z-score = -7.06, R) >droSec1.super_166 11448 67 - 36471 CCAUUUUGUGAUGCUC-------CAAAAUGCUCACCAAAAAGUGUAGCAUACAUAUUGGGGCAUCACA--AAAUGG ((((((((((((((((-------(((.((((((((......))).))))).....)))))))))))))--)))))) ( -36.30, z-score = -9.02, R) >droYak2.chr3R 3477270 67 + 28832112 CUAUUUUGUGGUGCUC-------CAAAAUGCUUACUGAAAAGUGUGGCCUACUUAUUGGGGCAUCACA--AAGUGG ((((((((((((((((-------(((...(((((((....)))).))).......)))))))))))))--)))))) ( -31.20, z-score = -6.51, R) >droEre2.scaffold_4770 10911939 67 - 17746568 CCAUUUUGUGGUGCUC-------UUAAAUGCUUACCGAAAAGUGUUGCCUACUUAUUAGGGCAUCGCA--CAAUGG (((((.((((((((((-------(..(((((((......)))))))...........)))))))))))--.))))) ( -23.02, z-score = -3.68, R) >droAna3.scaffold_12911 3159767 72 + 5364042 GCAUUUUGUGGCACCUCCAACCGUGAAGAGGUUUGCCAAAAAUAUUGCCUACC--UUUGAGCGCCACA--AAAUGG .((((((((((((((((..........)))))..((((((.............--)))).))))))))--))))). ( -24.52, z-score = -3.61, R) >dp4.chr2 14916023 60 + 30794189 CCAUUUUGCGCGGUCC-------------GGU---CCGAAAGUGUAGCAUACCUUUUUGGGCGCCACACAAAAUGG ((((((((.(.(((..-------------.((---(((((((.(((...))).)))))))))))).).)))))))) ( -22.80, z-score = -2.83, R) >consensus CCAUUUUGUGGUGCUC_______CAAAAUGCUUACCGAAAAGUGUAGCCUACUUAUUAGGGCAUCACA__AAAUGG ((((((((((((((((................(((......)))..............))))))))))..)))))) (-12.31 = -12.76 + 0.45)
| Location | 14,796,639 – 14,796,713 |
|---|---|
| Length | 74 |
| Sequences | 6 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.60810 |
| G+C content | 0.45336 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -3.62 |
| Energy contribution | -4.15 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.14 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
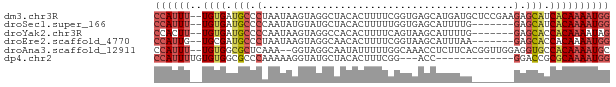
>dm3.chr3R 14796639 74 - 27905053 CCAUUU--UGUGAUGCCCUAAUAAGUAGGCUACACUUUUCGGUGAGCAUGAUGCUCCGAAGAGCAUCACAAAAUGG ((((((--((((((((((((.....))))......(((((((..(((.....)))))))))))))))))))))))) ( -32.80, z-score = -6.51, R) >droSec1.super_166 11448 67 + 36471 CCAUUU--UGUGAUGCCCCAAUAUGUAUGCUACACUUUUUGGUGAGCAUUUUG-------GAGCAUCACAAAAUGG ((((((--((((((((.((((.....(((((.((((....))))))))).)))-------).)))))))))))))) ( -35.50, z-score = -8.27, R) >droYak2.chr3R 3477270 67 - 28832112 CCACUU--UGUGAUGCCCCAAUAAGUAGGCCACACUUUUCAGUAAGCAUUUUG-------GAGCACCACAAAAUAG ....((--((((.(((.((((.......((...(((....)))..))...)))-------).))).)))))).... ( -14.80, z-score = -1.89, R) >droEre2.scaffold_4770 10911939 67 + 17746568 CCAUUG--UGCGAUGCCCUAAUAAGUAGGCAACACUUUUCGGUAAGCAUUUAA-------GAGCACCACAAAAUGG ((((((--((((((((((....((((.(....)))))...))...)))))...-------..)))).....))))) ( -14.20, z-score = -0.96, R) >droAna3.scaffold_12911 3159767 72 - 5364042 CCAUUU--UGUGGCGCUCAAA--GGUAGGCAAUAUUUUUGGCAAACCUCUUCACGGUUGGAGGUGCCACAAAAUGC .(((((--((((((((.((((--((((.....))))))))))..((((((........))))))))))))))))). ( -32.70, z-score = -5.46, R) >dp4.chr2 14916023 60 - 30794189 CCAUUUUGUGUGGCGCCCAAAAAGGUAUGCUACACUUUCGG---ACC-------------GGACCGCGCAAAAUGG (((((((((((((((..(..(((((((...))).))))..)---..)-------------)..))))))))))))) ( -21.20, z-score = -2.27, R) >consensus CCAUUU__UGUGAUGCCCAAAUAAGUAGGCUACACUUUUCGGUAAGCAUUUUG_______GAGCACCACAAAAUGG (((((...((((.(((..............................................))).)))).))))) ( -3.62 = -4.15 + 0.53)
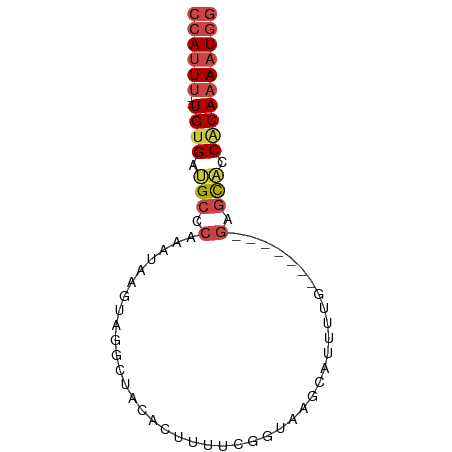
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:26 2011