| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,796,076 – 14,796,167 |
| Length | 91 |
| Max. P | 0.998835 |

| Location | 14,796,076 – 14,796,167 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.50 |
| Shannon entropy | 0.48086 |
| G+C content | 0.41037 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -13.74 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.998835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
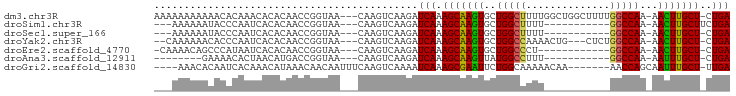
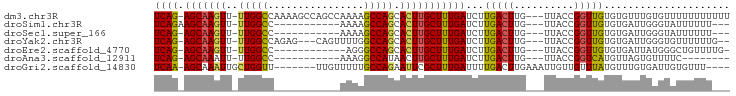
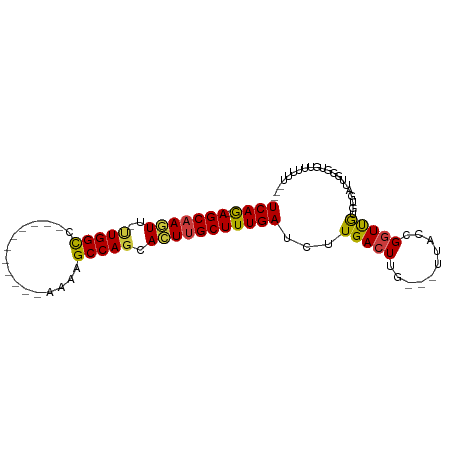
>dm3.chr3R 14796076 91 + 27905053 AAAAAAAAAAACACAAACACACAACCGGUAA---CAAGUCAAGAUCAAAGCAAGUGCUGGCUUUUGGCUGGCUUUUGGCCAA-AACUUGCU-CUGA ...............................---..........(((.(((((((..(((((...((....))...))))).-.)))))))-.))) ( -21.60, z-score = -0.96, R) >droSim1.chr3R 20843829 78 - 27517382 ---AAAAAAUACCCAAUCACACAACCGGUAA---CAAGUCAAGAUCAAAGCAAGUGCUGGCUUUU-----------GGCCAA-AACUUGCUUCUGA ---......((((.............)))).---..........(((((((((((..((((....-----------.)))).-.)))))))).))) ( -20.02, z-score = -2.55, R) >droSec1.super_166 10894 77 - 36471 ---AAAAAAUACCCAAUCACACAACCGGUAA---CAAGUCAAGAUCAAAGCAAGUGCUGGCUUUU-----------GGCCAA-AACUUGCU-CUGA ---......((((.............)))).---..........(((.(((((((..((((....-----------.)))).-.)))))))-.))) ( -20.32, z-score = -2.79, R) >droYak2.chr3R 3476728 86 + 28832112 --CAAAAAACACCCAAUCACACAACCGGUAA---CAAGUCAAGAUCAAAGCAAGUGCUGGCCAAAACUG---CUCUGGCCAA-AACUUGCU-CUGA --........(((.............)))..---..........(((.(((((((..((((((......---...)))))).-.)))))))-.))) ( -22.22, z-score = -2.92, R) >droEre2.scaffold_4770 10911397 78 - 17746568 -CAAAACAGCCCAUAAUCACACAACCGGUAA---CAAGUCAAGAUCAAAGCAAGUGCUGGCCCU------------GGCCAA-AACUUGCU-CUGA -.......(((...............)))..---..........(((.(((((((..((((...------------.)))).-.)))))))-.))) ( -20.16, z-score = -2.13, R) >droAna3.scaffold_12911 3159224 72 + 5364042 --------GAAAACACUAACAUGACCGGUAA---CAAGUCAAGAUCAAAGCAAGUUAUGGCCUUU-----------GGCCAA-AAUUUGCU-CUGA --------.............((((......---...))))...(((.((((((((.(((((...-----------))))).-))))))))-.))) ( -20.60, z-score = -3.22, R) >droGri2.scaffold_14830 1576089 84 - 6267026 ----AAACACAAUCACAAACAUAAACAACAAUUUCAAGUCAAAAUCAAAGCGAAUUCUGGCAAAAACAA-------AACCAGCAAUUUGCU-UUGA ----........................................((((((((((((((((.........-------..)))).))))))))-)))) ( -17.80, z-score = -5.29, R) >consensus ___AAAAAACACACAAUCACACAACCGGUAA___CAAGUCAAGAUCAAAGCAAGUGCUGGCCUUU___________GGCCAA_AACUUGCU_CUGA ................................................(((((((..(((((..............)))))...)))))))..... (-13.74 = -13.40 + -0.34)
| Location | 14,796,076 – 14,796,167 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 73.50 |
| Shannon entropy | 0.48086 |
| G+C content | 0.41037 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
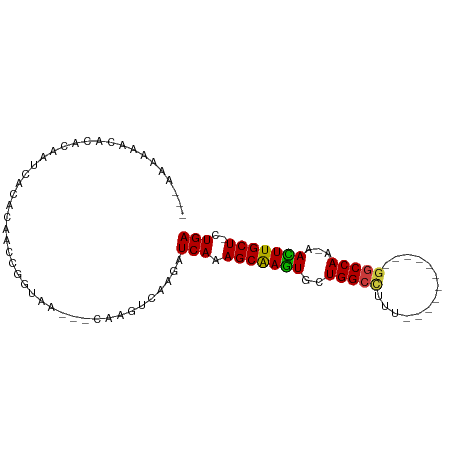
>dm3.chr3R 14796076 91 - 27905053 UCAG-AGCAAGUU-UUGGCCAAAAGCCAGCCAAAAGCCAGCACUUGCUUUGAUCUUGACUUG---UUACCGGUUGUGUGUUUGUGUUUUUUUUUUU ((((-(((((((.-(((((.....(....).....))))).)))))))))))....((((..---.....))))...................... ( -21.50, z-score = -0.33, R) >droSim1.chr3R 20843829 78 + 27517382 UCAGAAGCAAGUU-UUGGCC-----------AAAAGCCAGCACUUGCUUUGAUCUUGACUUG---UUACCGGUUGUGUGAUUGGGUAUUUUUU--- ...(((((((((.-(((((.-----------....))))).)))))))))............---...((((((....)))))).........--- ( -23.40, z-score = -2.01, R) >droSec1.super_166 10894 77 + 36471 UCAG-AGCAAGUU-UUGGCC-----------AAAAGCCAGCACUUGCUUUGAUCUUGACUUG---UUACCGGUUGUGUGAUUGGGUAUUUUUU--- ((((-(((((((.-(((((.-----------....))))).)))))))))))..........---...((((((....)))))).........--- ( -27.20, z-score = -3.39, R) >droYak2.chr3R 3476728 86 - 28832112 UCAG-AGCAAGUU-UUGGCCAGAG---CAGUUUUGGCCAGCACUUGCUUUGAUCUUGACUUG---UUACCGGUUGUGUGAUUGGGUGUUUUUUG-- ((((-(((((((.-((((((((((---...)))))))))).)))))))))))..........---...((((((....))))))..........-- ( -33.50, z-score = -3.90, R) >droEre2.scaffold_4770 10911397 78 + 17746568 UCAG-AGCAAGUU-UUGGCC------------AGGGCCAGCACUUGCUUUGAUCUUGACUUG---UUACCGGUUGUGUGAUUAUGGGCUGUUUUG- ((((-(((((((.-(((((.------------...))))).)))))))))))......((.(---(((((....).)))))...)).........- ( -26.40, z-score = -2.01, R) >droAna3.scaffold_12911 3159224 72 - 5364042 UCAG-AGCAAAUU-UUGGCC-----------AAAGGCCAUAACUUGCUUUGAUCUUGACUUG---UUACCGGUCAUGUUAGUGUUUUC-------- ((((-(((((.((-.(((((-----------...))))).)).)))))))))...(((((..---.....))))).............-------- ( -22.40, z-score = -3.21, R) >droGri2.scaffold_14830 1576089 84 + 6267026 UCAA-AGCAAAUUGCUGGUU-------UUGUUUUUGCCAGAAUUCGCUUUGAUUUUGACUUGAAAUUGUUGUUUAUGUUUGUGAUUGUGUUU---- ((((-(((.((((.(((((.-------........))))))))).)))))))....(((...((((....))))..))).............---- ( -17.20, z-score = -1.63, R) >consensus UCAG_AGCAAGUU_UUGGCC___________AAAAGCCAGCACUUGCUUUGAUCUUGACUUG___UUACCGGUUGUGUGAUUGGGUGUUUUUU___ ((((.((((((...(((((................)))))..))))))))))............................................ (-14.18 = -14.06 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:24 2011