
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,794,225 – 14,794,319 |
| Length | 94 |
| Max. P | 0.992522 |

| Location | 14,794,225 – 14,794,319 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.00 |
| Shannon entropy | 0.56762 |
| G+C content | 0.55799 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
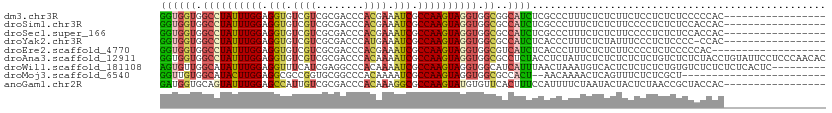
>dm3.chr3R 14794225 94 + 27905053 GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCACGAAAUCGCCAAGUAGGUGGCGGCAUCUCGCCCUUUCUCUCUUCUCCUCUCUCCCCCAC----------------- ((.((.(((((((((((.(((.((((........)))).))).))))))))))(((((....)))))..................).)).))..----------------- ( -31.80, z-score = -1.50, R) >droSim1.chr3R 20842001 94 - 27517382 GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCACGAAAUCGCCAAGUAGGUGGCGCCAUCUCGCCCUUUCUCUCUUCCCCUCUCUCCACCAC----------------- (((((.(((((((((((.(((.((((........)))).))).))))))))))((((......))))..................).)))))..----------------- ( -35.60, z-score = -3.23, R) >droSec1.super_166 9052 94 - 36471 GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCACGAAAUCGCCAAGUAGGUGGCGCCAUCUCGCCCUUUCUCUCUUCCCCUCUCUCCACCAC----------------- (((((.(((((((((((.(((.((((........)))).))).))))))))))((((......))))..................).)))))..----------------- ( -35.60, z-score = -3.23, R) >droYak2.chr3R 3474889 93 + 28832112 GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCAUGAAAUCGCCAAGUAGGUGGCGCCAUCUCACCCUUUCUCUAUUUCCCUCUCCCC-CCAC----------------- (((((((((((((((((.(((.((((........)))).))).))))))))))..)))))))...........................-....----------------- ( -28.60, z-score = -1.34, R) >droEre2.scaffold_4770 10909688 92 - 17746568 GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCACGAAAUCGCCAAGUAGGUGGCGUCAUCUCACCCUUUCUCUCUUCCCCUCUCCCCCAC------------------- ((((((.((((((((((.(((.((((........)))).))).)))))))))).))(....).)))).........................------------------- ( -27.50, z-score = -0.92, R) >droAna3.scaffold_12911 3157557 111 + 5364042 GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCACAAAAUCGCCAAGUAGGUGGCGCCUCUACCUCUAUUCUCUCUCUCUCUGUCUCUCUACCUGUAUUCCUCCCAACAC (((((.(((...((((..((.((((...)))))).))))...((((......))))))).))))).............................................. ( -26.80, z-score = -0.78, R) >droWil1.scaffold_181108 3061644 102 + 4707319 AGUGUUGGCAUAUUUGGAGGUUUCAUCGAGGCCCACAAAAUCGCCAAGUAGGUGGCAUCAUUUAACUAAAUGUCACUCUCUCUCUGUGUCUCUCUCUCACUC--------- (((((((((...((((..((((((...))))))..))))...)))))(..((((((((...........))))))))..).................)))).--------- ( -25.70, z-score = -2.04, R) >droMoj3.scaffold_6540 18772890 85 + 34148556 GGUUGUGGCAUACUUGGAGGCGCCGGUGCGGCCCACAAAAUCGCCAAGUAGGUGGCGCCACU--AACAAAACUCAGUUUCUCUCGCU------------------------ .((((((((....(((..((.((((...)))))).)))....((((......))))))))).--)))....................------------------------ ( -26.00, z-score = -0.01, R) >anoGam1.chr2R 58139997 94 - 62725911 GAUGGUGCAGUAUUUGGAGCCAUUGUCGCGACCCACAAAGGCGCCAAGUAUGUGUUCACUUUCCAUUUUCUAAUACUACUCUAACCGCUACCAC----------------- ..((((((.((((((((.(((.((((........)))).))).))))))))(((((...............)))))..........)).)))).----------------- ( -23.66, z-score = -1.87, R) >consensus GGUGGUGGCCUAUUUGGAGGUGUCGUCGCGACCCACGAAAUCGCCAAGUAGGUGGCGCCAUCUCACCCUUUCUCUCUUCCCCUCUCUCCCCCAC_________________ ((((((.((((((((((.(((.((((........)))).))).)))))))))).))..))))................................................. (-19.99 = -20.24 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:23 2011