
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,782,060 – 14,782,156 |
| Length | 96 |
| Max. P | 0.524318 |

| Location | 14,782,060 – 14,782,156 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Shannon entropy | 0.31956 |
| G+C content | 0.38651 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
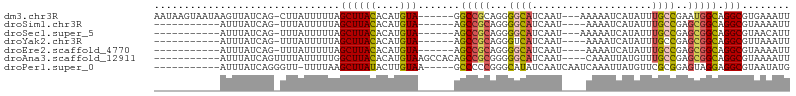
>dm3.chr3R 14782060 96 - 27905053 AAUAAGUAAUAAGUUAUCAG-CUUAUUUUUAGCUUACACAUGUA------GGCCGCAGGGGCAUCAAU---AAAAAUCAUAUUUGCCGAAUGGCAGGCGUGAAAUU ..((((.(((((((.....)-))))))))))((((((....)))------))).((....))......---.....(((..((((((....))))))..))).... ( -23.10, z-score = -1.73, R) >droSim1.chr3R 20827063 84 + 27517382 -----------AUUUAUCAG-UUUAUUUUUAGCUUACACAUGUA------AGCCGCAGGGGCAUCAAU----AAAAUCAUAUUUGCCGAGCGGCAGGCGUAAAAUU -----------.........-.....(((((((((((....)))------.(((((...((((..(((----(......))))))))..))))).))).))))).. ( -18.90, z-score = -1.59, R) >droSec1.super_5 751682 85 + 5866729 -----------AUUUAUCAG-UUUAUUUUUAGCUUACACAUGUA------AGCCGCAGGGGCAUCAAU---AAAAAUCAUAUUUGCCGAGCGGCAGGCGUAACAUU -----------.........-..........((((((....)))------.(((((...((((.....---............))))..))))).)))........ ( -17.83, z-score = -1.20, R) >droYak2.chr3R 3461392 84 - 28832112 -----------AUUUAUCAG-UUUAUUUUUAGCUUACACAUGUA------AGCCGCAGGGUCAUCAAU----AAAAUCAUAUUUGCCGAGCGGCAGGCGUUAAAUU -----------.........-......((((((((((....)))------)(((((..((.((..(((----(......))))))))..)))))....)))))).. ( -17.20, z-score = -1.50, R) >droEre2.scaffold_4770 10897497 84 + 17746568 -----------AUUUAUCAG-UUUAUUUUUAGCUUACACAUGUA------AGCCGCAGGGGCAUCAAU----AAAAUCAUAUUUGCCGAGCGGCAGGCGUAAAAUU -----------.........-.....(((((((((((....)))------.(((((...((((..(((----(......))))))))..))))).))).))))).. ( -18.90, z-score = -1.59, R) >droAna3.scaffold_12911 3145895 91 - 5364042 -----------AUUUAUCAGUUUUAUUUUUGGCUUACACAUGUAAGCCACAGCCGCGGGGGCAUCAAU----CAAAUUAUGUUUGCCGAGCGGCAGGCGUAAAAUU -----------..................((((((((....))))))))..(((.....)))......----....(((((((((((....))))))))))).... ( -30.70, z-score = -3.70, R) >droPer1.super_0 6314143 89 + 11822988 -----------AUUUAUCAGGGUU-UUUUAAGCUUAUACUUGUAA-----GCCCCCGGGCAUAUCAAUCAAUCAAAUUAUGUUCGCGGAGUAGGAGGCGUAAUAUG -----------..........(((-(((((.((((((....))))-----)).((((((((((..............)))))))).))..))))))))........ ( -19.74, z-score = -0.83, R) >consensus ___________AUUUAUCAG_UUUAUUUUUAGCUUACACAUGUA______AGCCGCAGGGGCAUCAAU____AAAAUCAUAUUUGCCGAGCGGCAGGCGUAAAAUU ...............................((((((....))........(((((...((((....................))))..)))))))))........ (-12.87 = -12.89 + 0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:20 2011