| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,746,118 – 14,746,211 |
| Length | 93 |
| Max. P | 0.581134 |

| Location | 14,746,118 – 14,746,211 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Shannon entropy | 0.36687 |
| G+C content | 0.49207 |
| Mean single sequence MFE | -15.98 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
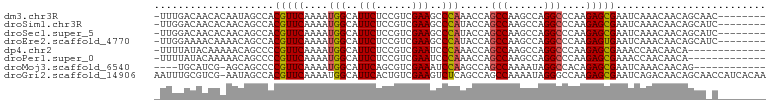
>dm3.chr3R 14746118 93 + 27905053 -UUUGACAACACAAUAGCCACGUUCAAAAUGGCAUUCUCCGUCGAAGCCCAAACCAGCCAAGCCAGGCCCAAGAGCGAAUCAAACAACAGCAAC-------- -...................(((((.....(((.(((......)))))).......(((......)))....))))).................-------- ( -15.70, z-score = -1.18, R) >droSim1.chr3R 20789515 93 - 27517382 -UUGGACAACACAACAGCCACGUUCAAAAUGGCAUUCUCCGUCGAAGCCCAUACCAGCCAAGCCAGGCCCAAGAGCGAAUCAAACAACAGCAUC-------- -.(((.(.........))))(((((.....(((.(((......)))))).......(((......)))....))))).................-------- ( -16.00, z-score = -0.57, R) >droSec1.super_5 715970 93 - 5866729 -UUGGACAACACAACAGCCACGUUCAAAAUGGCAUUCUCCGUCGAAGCCCAUACCAGCCAAGCCAGGCCCAAGAGCGAAUCAAACAACAGCAUC-------- -.(((.(.........))))(((((.....(((.(((......)))))).......(((......)))....))))).................-------- ( -16.00, z-score = -0.57, R) >droEre2.scaffold_4770 10861323 93 - 17746568 -UUGGAAAACAAAACAGCCACGUUCAAAAUGGCAUUCUCCGUCGAAGCCCAUACCAGCCAAGCCAGGCCCAAGAGUGAAUCAAACAACAGCAUC-------- -((((.........................(((.(((......)))))).......(((......)))))))......................-------- ( -12.30, z-score = 0.85, R) >dp4.chr2 14869711 88 + 30794189 -UUUUAUACAAAAACAGCCCCGUUCAAAAUGGCAUUCUCCGUCGAAUCCCAAACCAGCCAAGCCAGGCCCAAGAGCGAAACCAACAACA------------- -...................(((((....(((.((((......)))).))).....(((......)))....)))))............------------- ( -15.70, z-score = -2.74, R) >droPer1.super_0 6280837 88 - 11822988 -UUUUAUACAAAAACAGCCCCGUUCAAAAUGGCAUUCUCCGUCGAAUCCCAAACCAGCCAAGCCAGGCCCAAGAGCGAAACCAACAACA------------- -...................(((((....(((.((((......)))).))).....(((......)))....)))))............------------- ( -15.70, z-score = -2.74, R) >droMoj3.scaffold_6540 16632713 85 + 34148556 ----UGCAUCG-AGCAGCCCCGUUCAAAAUGGCAUUCAGCGUCGAAAUCCAAGCCAGCCAAAAUAGGCCACAGAGCGAAUCAAACAACAG------------ ----(((....-.)))....(((((....((((...................))))(((......)))....))))).............------------ ( -15.81, z-score = -0.83, R) >droGri2.scaffold_14906 955519 101 + 14172833 AAUUUGCGUCG-AAUAGCCACGUUCAAAAUGGCAUUCACUGUCGAAGUCUCAGCCAGCCAAAAUAGGGCCAAGAGCGAAUCAGACAACAGCAACCAUCACAA ...((((...(-(((.((((.........)))))))).((((((...(((..(((...........)))..))).)))..)))......))))......... ( -20.60, z-score = -0.79, R) >consensus _UUUGACAACAAAACAGCCACGUUCAAAAUGGCAUUCUCCGUCGAAGCCCAAACCAGCCAAGCCAGGCCCAAGAGCGAAUCAAACAACAGCA_C________ ....................(((((....(((..(((......)))..))).....(((......)))....)))))......................... (-12.15 = -12.19 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:17 2011