| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,744,119 – 14,744,215 |
| Length | 96 |
| Max. P | 0.999009 |

| Location | 14,744,119 – 14,744,215 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 58.37 |
| Shannon entropy | 0.84705 |
| G+C content | 0.43125 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.79 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
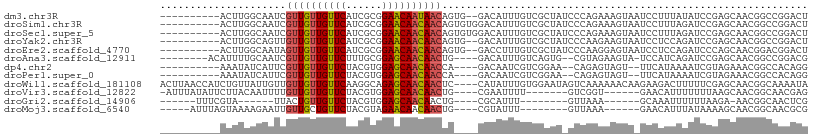
>dm3.chr3R 14744119 96 + 27905053 ----------ACUUGGCAAUCGUUGUUGUUCAUCGCGGAACAAUAACAGUG--GACAUUUGUCGCUAUCCCAGAAAGUAAUCCUUUAUAUCCGAGCAACGGCCGGACU ----------...((((....((((((((((......))))))))))....--(((....)))))))......................((((.((....)))))).. ( -25.80, z-score = -1.40, R) >droSim1.chr3R 20787515 98 - 27517382 ----------ACUUGGCAAUCGUUGUUGUUCAUCGCGGAACAACAACAGUGUGGACAUUUGUCGCUAUCCCAGAAAGUAAUCCUUUAGAUCCGAGCAACGGCCGGACU ----------...((((....((((((((((......))))))))))......(((....)))))))......................((((.((....)))))).. ( -27.70, z-score = -1.24, R) >droSec1.super_5 713974 98 - 5866729 ----------ACUUGGCAAUCGUUGUUGUUCAUCGCGGAACAACAACAGUGUGGACAUUUGUCGCUAUCCCAGAAAGUAAUCCUUUAGAUCCGAGCAACGGCCGGACU ----------...((((....((((((((((......))))))))))......(((....)))))))......................((((.((....)))))).. ( -27.70, z-score = -1.24, R) >droYak2.chr3R 3414059 96 + 28832112 ----------ACUUGGCAGUUGUUGUUGUUCAUCGCGGAACAACAACAGUG--GACAUUUGUCGCUAUCCCAAGAAGUAAUCCUCCAGAUCCGAGCAACGGCCGGACU ----------.(((((((.((((((((((((......))))))))))))))--(((....)))......)))))...............((((.((....)))))).. ( -31.50, z-score = -2.34, R) >droEre2.scaffold_4770 10859331 96 - 17746568 ----------ACUUGGCAAUAGUUGUUGUUCAUCGCGGAACAACAACAGUG--GACCUUUGUCGCUAUCCCAAGGAGUAAUCCUCCAGAUCCCAGCAACGGACGGACU ----------...........((((((((((......))))))))))....--..(((((((.((((((...((((....))))...)))...))).))))).))... ( -28.20, z-score = -1.52, R) >droAna3.scaffold_12911 3107665 93 + 5364042 --------ACAUUUUGCAAUCGUUGUUGUUCUUUGCGGAGCAACAACUG----GACAUUUGUCAGUG--CGUAGAAGUA-UCCAUCAGAUCCGAGCAACGGCCGGACG --------((.((((((....(((((((((((....)))))))))))..----(((....)))....--.)))))))).-.........((((.((....)))))).. ( -28.00, z-score = -1.54, R) >dp4.chr2 14867549 90 + 30794189 ----------AAAUAUCAUUCGUUGUUGUUCUACGUGGAGCAACAACCA----GACAAUCGUCGGAA--CAGAGUAGU--UUCAUAAAAUCGUAGAAACGGCCACAGG ----------.......(((((((((((((((....)))))))))))..----(((....)))....--..)))).((--(((...........)))))......... ( -21.80, z-score = -2.13, R) >droPer1.super_0 6278664 90 - 11822988 ----------AAAUAUCAUUCGUUGUUGUUCUACGUGGAGCAACAACCA----GACAAUCGUCGGAA--CAGAGUAGU--UUCAUAAAAUCGUAGAAACGGCCACAGG ----------.......(((((((((((((((....)))))))))))..----(((....)))....--..)))).((--(((...........)))))......... ( -21.80, z-score = -2.13, R) >droWil1.scaffold_181108 3000609 104 + 4707319 ACUUAACCAUCUGUUAUUGUUGUUGUUGUUCAAGGCAGAGCAACAACUC----CAUAUUUGUGGAAUAGUCAAAAAACAAGAAGACUUUUUCGAGCAACGGCAAAAUA ...........(((..(((((((((((((((......))))))))))..----........(((((.((((............)))).))))))))))..)))..... ( -26.20, z-score = -2.29, R) >droVir3.scaffold_12822 3309257 90 + 4096053 -AUUUAUAUUCUUACAAUUUUGUUGUUGUUCUACGUGGAGCAACAACUG----CGAAUUUU-------GUCGGU------GAACAUUUUUUUAAGCAACGGCAACGAG -..........((((......(((((((((((....)))))))))))..----(((.....-------.)))))------))................((....)).. ( -20.60, z-score = -1.68, R) >droGri2.scaffold_14906 953290 77 + 14172833 ------UUUCGUA------UUACUGUUGUUCUACGUGGAGCAACAACUG----CGCAUUU--------GUUAAA------GCAAAUUUUUUAAGA-AACGGCAACUCG ------.......------....(((((((((....)))))))))..((----(.(.(((--------.(((((------(......)))))).)-)).))))..... ( -16.10, z-score = -0.84, R) >droMoj3.scaffold_6540 16630229 85 + 34148556 -----AUUUAGUAAAAGAAUUGUUGCUGUUCUACGUAGAACAACAACUG----CGUAUUU--------GUUAAA------GAACAUUUAUAAAAGCAACGGCAACGCG -----................(((((((((((((((((........)))----)))...(--------(((...------.))))........)).)))))))))... ( -20.10, z-score = -1.92, R) >consensus __________ACUUGGCAAUCGUUGUUGUUCAACGCGGAACAACAACAG____GACAUUUGUCGCUA_CCCAAAAAGUA_UCCUUUAGAUCCGAGCAACGGCCAGACG .....................((((((((((......))))))))))............................................................. (-12.87 = -12.79 + -0.08)
| Location | 14,744,119 – 14,744,215 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 58.37 |
| Shannon entropy | 0.84705 |
| G+C content | 0.43125 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -8.45 |
| Energy contribution | -8.31 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
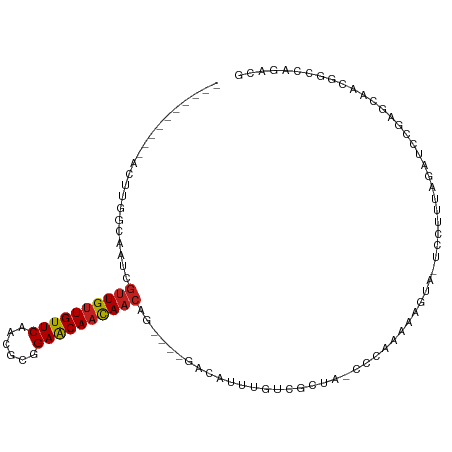
>dm3.chr3R 14744119 96 - 27905053 AGUCCGGCCGUUGCUCGGAUAUAAAGGAUUACUUUCUGGGAUAGCGACAAAUGUC--CACUGUUAUUGUUCCGCGAUGAACAACAACGAUUGCCAAGU---------- .....((((((((((((((....(((.....))))))))).))))).........--...((((.((((((......)))))).))))...)))....---------- ( -23.30, z-score = -0.22, R) >droSim1.chr3R 20787515 98 + 27517382 AGUCCGGCCGUUGCUCGGAUCUAAAGGAUUACUUUCUGGGAUAGCGACAAAUGUCCACACUGUUGUUGUUCCGCGAUGAACAACAACGAUUGCCAAGU---------- .....(((.((((((...((((...(((......))).))))))))))............(((((((((((......)))))))))))...)))....---------- ( -30.10, z-score = -1.76, R) >droSec1.super_5 713974 98 + 5866729 AGUCCGGCCGUUGCUCGGAUCUAAAGGAUUACUUUCUGGGAUAGCGACAAAUGUCCACACUGUUGUUGUUCCGCGAUGAACAACAACGAUUGCCAAGU---------- .....(((.((((((...((((...(((......))).))))))))))............(((((((((((......)))))))))))...)))....---------- ( -30.10, z-score = -1.76, R) >droYak2.chr3R 3414059 96 - 28832112 AGUCCGGCCGUUGCUCGGAUCUGGAGGAUUACUUCUUGGGAUAGCGACAAAUGUC--CACUGUUGUUGUUCCGCGAUGAACAACAACAACUGCCAAGU---------- .....(((.((((((...((((.(((((....))))).)))))))))).......--...(((((((((((......)))))))))))...)))....---------- ( -33.80, z-score = -2.51, R) >droEre2.scaffold_4770 10859331 96 + 17746568 AGUCCGUCCGUUGCUGGGAUCUGGAGGAUUACUCCUUGGGAUAGCGACAAAGGUC--CACUGUUGUUGUUCCGCGAUGAACAACAACUAUUGCCAAGU---------- .........((((((...((((.(((((....))))).))))))))))...(((.--....((((((((((......))))))))))....)))....---------- ( -34.10, z-score = -2.38, R) >droAna3.scaffold_12911 3107665 93 - 5364042 CGUCCGGCCGUUGCUCGGAUCUGAUGGA-UACUUCUACG--CACUGACAAAUGUC----CAGUUGUUGCUCCGCAAAGAACAACAACGAUUGCAAAAUGU-------- .(((((((....)).)))))....((((-....)))).(--((..(((....)))----..(((((((.((......)).)))))))...))).......-------- ( -22.70, z-score = -0.44, R) >dp4.chr2 14867549 90 - 30794189 CCUGUGGCCGUUUCUACGAUUUUAUGAA--ACUACUCUG--UUCCGACGAUUGUC----UGGUUGUUGCUCCACGUAGAACAACAACGAAUGAUAUUU---------- .........(((((...........)))--))......(--(((.(((....)))----..(((((((.((......)).))))))))))).......---------- ( -15.50, z-score = 0.29, R) >droPer1.super_0 6278664 90 + 11822988 CCUGUGGCCGUUUCUACGAUUUUAUGAA--ACUACUCUG--UUCCGACGAUUGUC----UGGUUGUUGCUCCACGUAGAACAACAACGAAUGAUAUUU---------- .........(((((...........)))--))......(--(((.(((....)))----..(((((((.((......)).))))))))))).......---------- ( -15.50, z-score = 0.29, R) >droWil1.scaffold_181108 3000609 104 - 4707319 UAUUUUGCCGUUGCUCGAAAAAGUCUUCUUGUUUUUUGACUAUUCCACAAAUAUG----GAGUUGUUGCUCUGCCUUGAACAACAACAACAAUAACAGAUGGUUAAGU .((((.((((((....(((......))).((((..(((.....((((......))----))(((((((.((......)).)))))))..))).)))))))))).)))) ( -22.80, z-score = -1.41, R) >droVir3.scaffold_12822 3309257 90 - 4096053 CUCGUUGCCGUUGCUUAAAAAAAUGUUC------ACCGAC-------AAAAUUCG----CAGUUGUUGCUCCACGUAGAACAACAACAAAAUUGUAAGAAUAUAAAU- ......................((((((------..(((.-------.....)))----..(((((((.((......)).)))))))..........))))))....- ( -14.10, z-score = -0.58, R) >droGri2.scaffold_14906 953290 77 - 14172833 CGAGUUGCCGUU-UCUUAAAAAAUUUGC------UUUAAC--------AAAUGCG----CAGUUGUUGCUCCACGUAGAACAACAGUAA------UACGAAA------ .(((((((((((-(.(((((........------))))).--------))))).)----))......))))..((((..((....))..------))))...------ ( -13.60, z-score = -0.03, R) >droMoj3.scaffold_6540 16630229 85 - 34148556 CGCGUUGCCGUUGCUUUUAUAAAUGUUC------UUUAAC--------AAAUACG----CAGUUGUUGUUCUACGUAGAACAGCAACAAUUCUUUUACUAAAU----- .((((.((....)).........((((.------...)))--------)...)))----).(((((((((((....)))))))))))................----- ( -20.00, z-score = -2.52, R) >consensus CGUCCGGCCGUUGCUCGGAUAUAAAGGA_UACUUUUUGGG_UAGCGACAAAUGUC____CAGUUGUUGCUCCGCGAAGAACAACAACGAUUGCCAAAU__________ .............................................................(((((((.((......)).)))))))..................... ( -8.45 = -8.31 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:15 2011