
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,740,988 – 14,741,087 |
| Length | 99 |
| Max. P | 0.653674 |

| Location | 14,740,988 – 14,741,087 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 59.52 |
| Shannon entropy | 0.81512 |
| G+C content | 0.35988 |
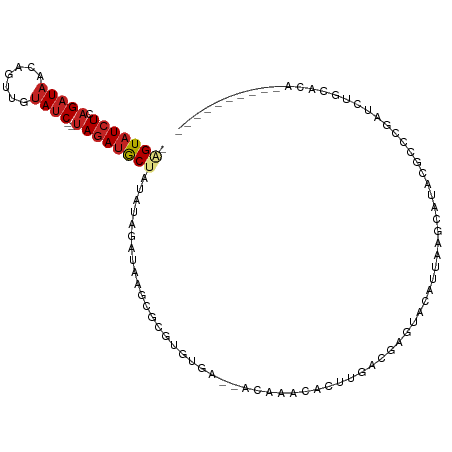
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -5.60 |
| Energy contribution | -5.76 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.653674 |
| Prediction | RNA |
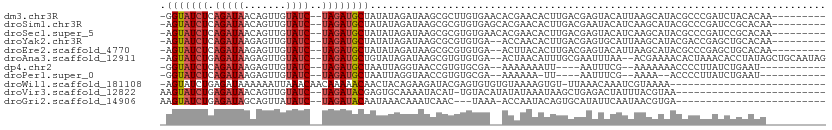
Download alignment: ClustalW | MAF
>dm3.chr3R 14740988 99 + 27905053 -GGUAUCUCAGAUAACAGUUGUAUC--UAGAUGCUAUAUAGAUAAGCGCUUGUGAACACGAACACUUGACGAGUACAUUAAGCAUACGCCCGAUCUACACAA--------- -(((((((.(((((.(....)))))--))))))))...(((((..(((((((((.((.((.........)).)).)).)))))....))...))))).....--------- ( -20.40, z-score = -0.74, R) >droSim1.chr3R 20784481 99 - 27517382 -AGUAUCUCAGAUAACAGUUGUAUC--UAGAUGCUAUAUAGAUAAGCGCGUGUGAGCACGAACACUUGACGAAUACAUCAAGCAUACGCCCGAUCCGCACAA--------- -(((((((.(((((.(....)))))--)))))))).....(((..(.(((((((.(......).(((((........)))))))))))))..))).......--------- ( -23.20, z-score = -1.13, R) >droSec1.super_5 711075 99 - 5866729 -AGUAUCUCAGAUAACAGUUGUAUC--UAGAUGCUAUAUAGAUAAGCGCGUGUGAACACGAACACUUGACGAGUACAUCAAGCAUACGCCCGAUCCGCACAA--------- -(((((((.(((((.(....)))))--)))))))).....(((..(((((((....))))....(((((........)))))....)))...))).......--------- ( -23.80, z-score = -1.36, R) >droYak2.chr3R 3411137 97 + 28832112 -AGUAUCUCAGAUAAGAGUUGUAUC--UAGAUGCUAUAUAGAUAAGCGCGUGUGA--ACCAACACUUGACGAGUGCAUUAAGCAUACGACCGAGCUGCACAA--------- -(((((((.(((((.......))))--))))))))..........((((((((..--....))))....((.((((.....)))).)).....)).))....--------- ( -24.50, z-score = -1.23, R) >droEre2.scaffold_4770 10856398 97 - 17746568 -AGUAUCUCAGAUAAGAGUUGUAUC--UAGAUGCUAUAUAGAUAAGCGCGUGUGA--ACUUACACUUGACGAGUACAUUAAGCAUACGCCCGAGCUGCACAA--------- -(((((((.(((((.......))))--)))))))).....(...((((((((((.--......((((...))))........)))))))....))).)....--------- ( -21.66, z-score = -0.23, R) >droAna3.scaffold_12911 3104211 104 + 5364042 -AGUAUCUGAGAUAAGAGUUGUAUC--UAGAUGCUGUAUAGAUAAGCGUGUGUGA--ACUAACAUUUGCGAAUUUAA--ACGAAAACACUAAACACCUAUAGCUGCAAUAG -(((((((.(((((.......))))--))))))))((.(((((..(((.((((..--....)))).)))..))))).--)).............................. ( -20.50, z-score = -0.76, R) >dp4.chr2 14864117 89 + 30794189 -GGUAUCUCAGAUAAGAGUUGUAUC--UAGAUGCUAAUUAGGUAACCGUGUGCGA--AAAAAAAUU----AAUUUCG--AAAAAAACCCCUUAUCUGAAU----------- -(((((((.(((((.......))))--))))))))..((((((((..((...(((--((.......----..)))))--......))...))))))))..----------- ( -17.60, z-score = -2.54, R) >droPer1.super_0 6275630 86 - 11822988 -GGUAUCUCAGAUAAGAGUUGUAUC--UAGAUGCUAAUUAGGUAACCGUGUGCGA--AAAAAA-UU----AAUUUCG--AAAA--ACCCCUUAUCUGAAU----------- -(((((((.(((((.......))))--))))))))..((((((((..(.((.(((--((....-..----..)))))--....--)))..))))))))..----------- ( -19.60, z-score = -3.11, R) >droWil1.scaffold_181108 2996682 83 + 4707319 -AGUAUCUGAGAUAAAAAAUUAAACAACAAAAACAACUACAGAAGAUACGAGUGUGUGUAAAAGUGU-UUAAACAAAUCGUAAAA-------------------------- -....((((((.........................)).))))...(((((...(((.((((....)-))).)))..)))))...-------------------------- ( -9.31, z-score = -0.17, R) >droVir3.scaffold_12822 3305805 83 + 4096053 AAGUAUCUGAGAUAACAGUUGUAUC--UAGAUACGAGUGCAAAAUACAU-UGUACAUAUAUAAAUAAGCUGAGACUAUUUACGUAA------------------------- ..((((((.(((((.(....)))))--)))))))..((((((......)-))))).((..((((((.........))))))..)).------------------------- ( -16.90, z-score = -1.57, R) >droGri2.scaffold_14906 950095 80 + 14172833 AAGUAUCUGAGAUAGCAGUUAUAUC--UAGAUACAAUAAACAAAUCAAC---UAAA-ACCAAUACAGUGCAUAUUCAAUAACGUGA------------------------- ..((((((.(((((.......))))--)))))))...............---....-.............................------------------------- ( -10.60, z-score = -1.18, R) >consensus _AGUAUCUCAGAUAACAGUUGUAUC__UAGAUGCUAUAUAGAUAAGCGCGUGUGA__ACAAACACUUGACGAGUACAUUAAGCAUACGCCCGAUCUGCACA__________ ..((((((..((((.......))))...))))))............................................................................. ( -5.60 = -5.76 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:14 2011