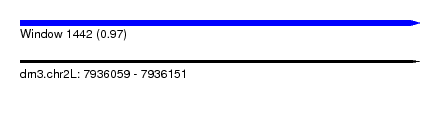
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,936,059 – 7,936,151 |
| Length | 92 |
| Max. P | 0.971703 |

| Location | 7,936,059 – 7,936,151 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 58.70 |
| Shannon entropy | 0.73035 |
| G+C content | 0.50421 |
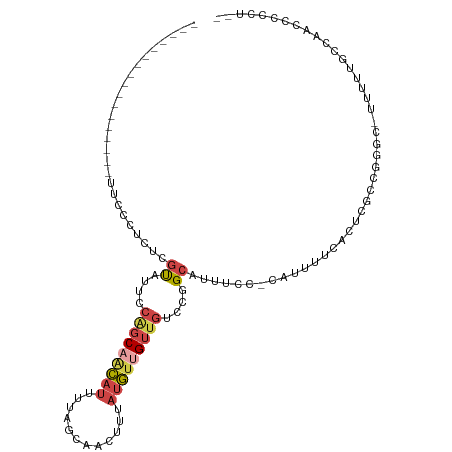
| Mean single sequence MFE | -17.77 |
| Consensus MFE | -5.49 |
| Energy contribution | -6.27 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
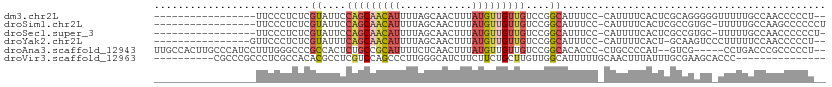
>dm3.chr2L 7936059 92 + 23011544 -----------------UUCCCUCUCGUAUUCCAGCAACAUUUUAGCAACUUUAUGUUGUUGUCCGGCAUUUCC-CAUUUUCACUCGCAGGGGGUUUUUGCCAACCCCCU-- -----------------.........((....(((((((((............)))))))))....))......-.............((((((((......))))))))-- ( -23.20, z-score = -3.11, R) >droSim1.chr2L 7731941 93 + 22036055 -----------------UUCCCUCUCGUAUUCCAGCAACAUUUUAGCAACUUUAUGUUGUUGUCCGGCAUUUCC-CAUUUUCACUCGCCGUGC-UUUUUGCCAAGCCCCCCU -----------------.........(((....((((......(((((((.....)))))))..((((......-...........)))))))-)...)))........... ( -15.13, z-score = -2.54, R) >droSec1.super_3 3450168 92 + 7220098 -----------------UUCCCUCUCGUAUUCCAGCAACAUUUUAGCAACUUUAUGUUGUUGUCCGGCAUUUCC-CAUUUUCACUCGCCGUGC-UUUUUGCCAACCCCCCU- -----------------.........(((....((((......(((((((.....)))))))..((((......-...........)))))))-)...)))..........- ( -15.13, z-score = -3.13, R) >droYak2.chr2L 17366351 92 - 22324452 ----------------GUUCCCUCUCGUAUUUCAGCAACAUUUUAGCAACUUUAUGUUGUUGUCCGGCAUUUCC-CAUUUUCACU-GCAAGUCCCUUUUUCCAACCCCCU-- ----------------..........((....(((((((((............)))))))))....))......-..........-........................-- ( -11.00, z-score = -2.35, R) >droAna3.scaffold_12943 3667735 102 - 5039921 UUGCCACUUGCCCAUCCUUUGGGCCCGCCACUCUGCCGCAUUUUCUCAACUUUAUGUUGUUGUCCGGCACACCC-CUGCCCCAU--GUCG-----CCUGACCCGCCCCCU-- .........(((((.....)))))..((...((.((.((((.....((((.....))))......((((.....-.))))..))--)).)-----)..))...)).....-- ( -21.60, z-score = -1.43, R) >droVir3.scaffold_12963 8710344 87 + 20206255 ----------CGCCCGCCCUCGCCACACGCCUCGUCCAGCCCUUGGGCAUCUUCUUCUGCUUGUUGGCAUUUUUGCAACUUUAUUUGCGAAGCACCC--------------- ----------.....((....((((((.((........(((....)))..........)).)).))))...(((((((......)))))))))....--------------- ( -20.57, z-score = -0.69, R) >consensus _________________UUCCCUCUCGUAUUCCAGCAACAUUUUAGCAACUUUAUGUUGUUGUCCGGCAUUUCC_CAUUUUCACUCGCCGGGC_UUUUUGCCAACCCCCU__ ..........................((....(((((((((............)))))))))....))............................................ ( -5.49 = -6.27 + 0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:44 2011