| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,729,629 – 14,729,734 |
| Length | 105 |
| Max. P | 0.951258 |

| Location | 14,729,629 – 14,729,734 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 60.39 |
| Shannon entropy | 0.81671 |
| G+C content | 0.51074 |
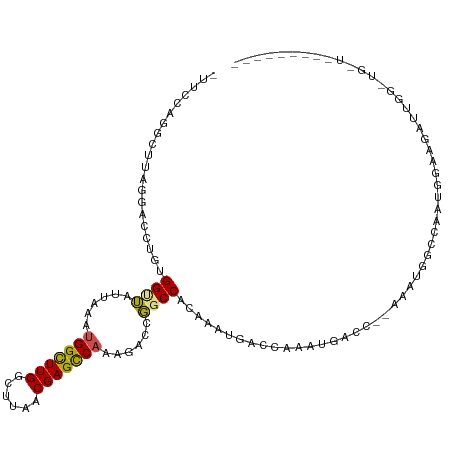
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -13.23 |
| Energy contribution | -12.91 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
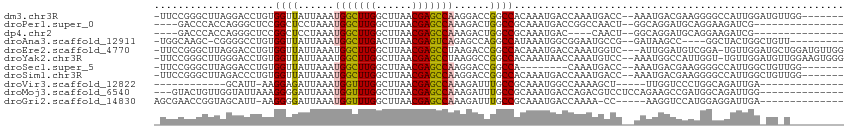
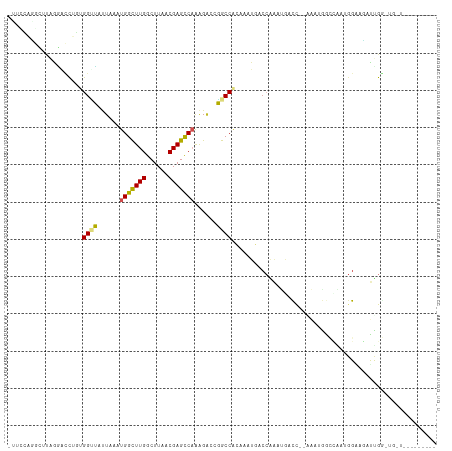
>dm3.chr3R 14729629 105 - 27905053 -UUCCGGGCUUAGGACCUGUGGUUAUUAAAUGGCUUGGCUUAACGAGCCAAGGACCGGCCACAAAUGACCAAAUGACC--AAAUGACGAAGGGGCCAUUGGAUGUUGG------- -.(((.((((..((.....((((((((...(((((((......))))))).((.....))...)))))))).....))--......(....)))))...)))......------- ( -31.90, z-score = -0.84, R) >droPer1.super_0 6262572 94 + 11822988 ----GACCCACCAGGGCUCCGGCUCCUAAAUGGCUUGGCUUAACGAGCCAAAGACUGGCCGCAAAUGACCGGCCAACU--GGCAGGAUGCAGGAAGAUCG--------------- ----..(((....))).(((.((((((...(((((((......))))))).....((((((........))))))...--...)))).)).)))......--------------- ( -35.70, z-score = -1.96, R) >dp4.chr2 14851185 90 - 30794189 ----GACCCACCAGGGCUCCGGCUCCUAAAUGGCUUGGCUUAACGAGCCAAAGACUGGCCGCAAAUGAC----CAACU--GGCAGGAUGCAGGAAGAUCG--------------- ----..(((....))).(((((((......(((((((......)))))))......))))(((..((.(----(....--))))...))).)))......--------------- ( -30.20, z-score = -1.25, R) >droAna3.scaffold_12911 3092034 98 - 5364042 -UGGCAAGC-CGGGGCCUGUGGUUAUUAAAUGGCUUGACUUAACGAGUCAGAGCCAGGCCAUAAAUGGCGGAAUGCCG--GAUAAGCC----GGCUACUGGCUGUU--------- -.((((.((-(((.((((((((((......(((((((((((...)))))).))))))))))))...))).....((((--(.....))----)))..)))))))))--------- ( -41.80, z-score = -2.82, R) >droEre2.scaffold_4770 10844689 110 + 17746568 -UUCCGGGCUUAGGACCUGUGGUUAUUAAAUGGCUUGGCUUAACGAGCCUAAGACCGGCCACAAAUGACCAAAUGGUC---AUUGGAUGUCGGA-UGUUGGAUGCUGGAUGUUGG -.(((((.((((.((((...)))).......((((((......)))))))))).(((((....(((((((....))))---)))....))))).-.........)))))...... ( -36.80, z-score = -1.90, R) >droYak2.chr3R 3399675 111 - 28832112 -UUCCGGGCUUGGGACCUGUGGUUAUUAAAUGGCUUGGCUUAACGAGCCUAAGGCCGGCCACAAAUAACCAAAUGUCC--AAAUGGCCAUUGGU-UGUUGGAUGUUGGAAGUGGG -..((((.((((.((((...)))).......((((((......)))))))))).))))((((......((((.(((((--(.(..(((...)))-..)))))))))))..)))). ( -40.10, z-score = -1.64, R) >droSec1.super_5 699738 97 + 5866729 -UUCCGGGCUUAGGACCUGUGGUUAUUAAAUGGCUUGGCUUAACGAGCCAAGGACCGGCCA--------CAAAUGACC--AAAUGACGAAGGGGCCAUUGGCUGUUGG------- -..(((((((..((.(((.((((((((...(((((((......))))))).((.....)).--------..)))))))--)........)))..))...))))..)))------- ( -29.90, z-score = -0.34, R) >droSim1.chr3R 20772470 105 + 27517382 -UUCCGGGCUUAGACCCUGUGGUUAUUAAAUGGCUUGGCUUAACGAGCCAAGGACCGGCCACAAAUGACCAAAUGACC--AAAUGACGAAGGGGCCAUUGGCUGUUGG------- -....(((......)))..((((((((...(((((((......))))))).((.....))...)))))))).....((--((....(....)((((...)))).))))------- ( -30.90, z-score = -0.27, R) >droVir3.scaffold_12822 3282134 83 - 4096053 ------------GCAUU-AAGGAGAUUAAAUGGUUUGGCUUAACGAGCCAAAGAUUUGCCGCAAAUGGCCAAAAGCU-----UUGGUCCCUGGCAGAUUGA-------------- ------------.....-............(((((((......)))))))..(((((((((.....((((((.....-----))))))..)))))))))..-------------- ( -24.40, z-score = -1.14, R) >droMoj3.scaffold_6540 19182325 98 + 34148556 ---GUACUGUUGGUAUUAAAGGGGAUUAAAUGGUUUGGCUUAACGAGCCAAAGAUUUGCCGCAAAUGACCAGACGUCCUCCAGAAGCCGAUGGCAGAUUGG-------------- ---...((((((((.((...((((((....(((((((((((...))))))...(((((...))))))))))...))))))..)).))))))))........-------------- ( -27.40, z-score = -1.13, R) >droGri2.scaffold_14830 2681154 94 + 6267026 AGCGAACCGGUAGCAUU-AAGGGGAUUAAAUGGUUUGGCUUAACGAGCCAAAGAUUUGCCGCAAAUGACCAAAA-CC-----AAGGUCCAUGGAGGAUUGA-------------- ..(((.(((((..((((-..((.((.....(((((((......))))))).....)).))...)))))))....-((-----(.......))).)).))).-------------- ( -21.90, z-score = 0.06, R) >consensus _UUCCAGGCUUAGGACCUGUGGUUAUUAAAUGGCUUGGCUUAACGAGCCAAAGACCGGCCACAAAUGACCAAAUGACC__AAAUGGCCAAUGGAAGAUUGG_UG_U_________ ....................((((......(((((((......)))))))......))))....................................................... (-13.23 = -12.91 + -0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:12 2011