
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,725,207 – 14,725,295 |
| Length | 88 |
| Max. P | 0.598675 |

| Location | 14,725,207 – 14,725,295 |
|---|---|
| Length | 88 |
| Sequences | 12 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.65259 |
| G+C content | 0.62141 |
| Mean single sequence MFE | -34.29 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14725207 88 - 27905053 GUCCAGCACGGAACCAUUGCCACUCGACUGGGCCUGAAACCUGGGCCCAGAAU--GGGCCAGG---UCGAAUGGC----GUCUCCGGCAAAUGACUG (((..((.((((.....(((((.((((((.((((((....(((....)))..)--))))).))---)))).))))----)..))))))....))).. ( -37.90, z-score = -2.63, R) >droSim1.chr3R 20768017 88 + 27517382 GUCCAGCACGGAACCAUUGCCGCUCGACUGGGCCUGAAACCUGGGCCCAGAAA--GGGCCAGG---UCGAAUGGC----GUCACCGGCAAAUGACUG (((..((.(((......(((((.((((((.(((((.....(((....)))...--))))).))---)))).))))----)...)))))....))).. ( -36.40, z-score = -1.91, R) >droSec1.super_5 695354 88 + 5866729 GUCCAGCACGGAACCAUUGCCGCUCGACUGGGCCUGAAACCUGGGCCCAGAAU--GGGCCAGG---UCGAAUGGC----GUCUCCGGCAAAUGACUG (((..((.((((.....(((((.((((((.((((((....(((....)))..)--))))).))---)))).))))----)..))))))....))).. ( -37.20, z-score = -2.06, R) >droYak2.chr3R 3394352 88 - 28832112 GUCCAGCACGGAACCAUUGCCGCUCGACUGGGCCUGGAACCUGGGCCCAGAAA--GGGCCAGG---UCGAAUGGC----GUCUCUGGCAAAUGACUG (((..((.((((.....(((((.((((((.(((((.....(((....)))...--))))).))---)))).))))----)..))))))....))).. ( -36.10, z-score = -1.44, R) >droEre2.scaffold_4770 10836587 88 + 17746568 GUCCAGCACGGAACCAUUGCCGCUCGACUGGGCCUGAAACCUGGGCCCAGAAA--GGGCCAAG---UCCAAUGGC----GUCUCCGGCAAGUGAGUG .....((.((((.((((((..((((..((((((((.......))))))))...--))))....---..)))))).----...))))))......... ( -34.30, z-score = -1.12, R) >droAna3.scaffold_12911 3086315 88 - 5364042 GUCCAGCACGGAUCCAUUGCCGCCCGACUGGGCCUGGAACCUGGGCCCUGUGA--GGGCCAAA---GUCAAUGGC----GUCUCCGGAAAAUGAGUG ........((((.(((((((.((((.((.(((((..(...)..))))).))..--))))....---).)))))).----...))))........... ( -33.70, z-score = -0.75, R) >dp4.chr2 14847596 88 - 30794189 GUCCAGCACGGAUCGAUUGCCUCCUGUUUGGGCCUCGAAUCUCGGCCCCAG--GAGUGCCAUG---GCCAACGGC----GGCUCUGGCAACAGACUG ((((.....)))).......((((((...(((((.........))))))))--)))(((((.(---(((......----)))).)))))........ ( -35.20, z-score = -1.18, R) >droPer1.super_0 6258889 88 + 11822988 GUCCAGCACGGAACGAUUGCCUCCUGUUUGGGCCUCGAACCUGGGCCCCAA--GAGUGCCGUG---GCCAACGGC----GGCUCUGGCAACAGACUG .(((.....)))......(((..((.((.((((((.......)))))).))--.)).(((((.---....)))))----)))((((....))))... ( -34.40, z-score = -0.67, R) >droWil1.scaffold_181108 2979418 73 - 4707319 GUCCAGCACGGAGCCAUUGGCACCCGACUGUGCCUCAAAUCUGGGCCCAGACA--UGGCCAUG---GUCAAUGGC----GUC--------------- .(((.....)))(((((((((......(((.(((.((....))))).))).((--(....)))---)))))))))----...--------------- ( -27.10, z-score = -0.80, R) >droMoj3.scaffold_6540 19177193 97 + 34148556 AUCCAGGACGGAGCCAUUGCCACUCGCCUGGGCAUCGAAUCUGGGCCCAAGUCCACUGACAGGCCAGGCAGCGUCCAAUGUCUGCGGCAACAAGCUG .(((.....)))((..(((((.((.((((((((.(((....)))))))..(((....))))))).))(((((((...))).)))))))))...)).. ( -33.60, z-score = 0.11, R) >droGri2.scaffold_14830 2676154 97 + 6267026 GUCCAGCACGGAGCCAUUACCGCCCGCCUGGGCCUCGAAUCGCGGCCCAAGUUGAGUGCCAAACAUGGCAACCGCGAUUGUCUCUGGCAUCAAACUG .....((.(((((.((....(((..((.((((((.((...)).)))))).)).(..(((((....)))))..))))..)).)))))))......... ( -33.90, z-score = -1.35, R) >anoGam1.chr2R 21814003 88 - 62725911 GUCCACCACGGUCAGGUUGGCCACCGGCUGGCCCCACAGCCCGGGCUGUUGCAUUGGACCGUA---GU--AUGGC----AUCUUUGAAGAAUUUCUG (((.((.((((((.(((..(((...)))..)))..(((((....))))).......)))))).---))--..)))----.................. ( -31.70, z-score = -0.71, R) >consensus GUCCAGCACGGAACCAUUGCCGCCCGACUGGGCCUGGAACCUGGGCCCAGAAA__GGGCCAGG___GCCAAUGGC____GUCUCCGGCAAAUGACUG .(((.....))).............(((.((((((.......)))))).........((((..........))))....)))............... (-13.28 = -13.70 + 0.42)

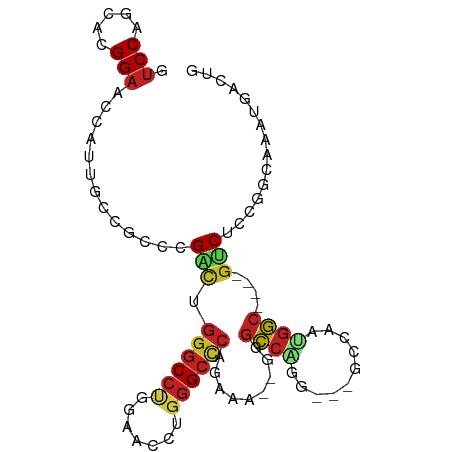

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:11 2011