
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,717,366 – 14,717,476 |
| Length | 110 |
| Max. P | 0.710242 |

| Location | 14,717,366 – 14,717,476 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.63 |
| Shannon entropy | 0.56498 |
| G+C content | 0.46244 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.710242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14717366 110 - 27905053 ------AUCCGAAACGUAUGUAUUCGCAUCGCCCACUUUGCGUCAUUCGGCGUUCGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACACCAAGCAGU ------...((((.(((.......((((..........)))).......))))))).....(((((((((.(((((......))))))))).........(....).....))))) ( -25.24, z-score = -1.13, R) >droSim1.chr3R 20759684 109 + 27517382 ------AUCCCAAACGUAUGUAUUCGCAACGCCCACUCUGCGUCAUUCGGCGGGCGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACACCAAGCAG- ------...................((..(((((.((...........)).)))))..((((((..((((.(((((......))))))))).....)))))).........))..- ( -26.20, z-score = -1.03, R) >droSec1.super_5 687541 109 + 5866729 ------AUCCCAAACGUAUGUAUUCGCAACGCCCACUCUGCGUCAUUCGGCGGGCGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACACCAAGCAG- ------...................((..(((((.((...........)).)))))..((((((..((((.(((((......))))))))).....)))))).........))..- ( -26.20, z-score = -1.03, R) >droYak2.chr3R 3385668 114 - 28832112 CUCUCAAUCUGAAACGUAUGUAUUCGCAUCG-CCACUCUGCGUCAUUCGACGGACGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACACCAAGCAG- ..............((.((((....))))))-.....(((((((....)))((.....((((((..((((.(((((......))))))))).....)))))).....))..))))- ( -25.50, z-score = -1.01, R) >droEre2.scaffold_4770 10829026 109 + 17746568 ------AUCUGAAACGUAUGUAUUCGCAUCGCCCACUCUGCGUCAUUCGGCUGGCGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACACCAAGCGG- ------........(((.((..(((((...(((...............)))..)))))((((((..((((.(((((......))))))))).....))))))......)).))).- ( -26.06, z-score = -0.40, R) >droAna3.scaffold_12911 3078398 86 - 5364042 -----------------------------CGCCCACACUUCGUCAUUUGGCGAAUGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACAUGAAACUG- -----------------------------......((.((((((....))))))))..((((((..((((.(((((......))))))))).....)))))).............- ( -20.40, z-score = -1.70, R) >droWil1.scaffold_181108 2966979 100 - 4707319 ----------------AUCCCCGGCCCUUCUAAUAUCAUGCCCUUUUUUACGAAUGAAAUUGCUUUGCUCCAAGUGUAAUAUCACUUAAGCUUUUAAGCAGUCAAACCCAGAGUAC ----------------......((...........((((..............)))).(((((((.(((..(((((......))))).)))....))))))).....))....... ( -15.64, z-score = -1.10, R) >droGri2.scaffold_14830 2667468 110 + 6267026 ---UCGGUUCCCUAGAAUUCUCGUUGGACCGCGCACUGCGC---AUUUUGUGGAUGAAAUCGCUGCGCUCCAAGUGUAAUUUCACUUAAGCUCUUUAGCAAACAGCACCGCCCUGC ---..(((((....((....))...)))))((((...))))---.....((((..(....)((((......(((((......)))))..(((....)))...)))).))))..... ( -26.30, z-score = 0.37, R) >consensus ______AUCCCAAACGUAUGUAUUCGCAUCGCCCACUCUGCGUCAUUCGGCGGACGAAAUUGCUGCGCUUCAAGUGUAAUAUCACUUAAGCUCUUAAGCAGUCAACACCAAGCAG_ .........................((..((((...............))))......((((((..((((.(((((......))))))))).....)))))).........))... (-13.76 = -14.20 + 0.44)
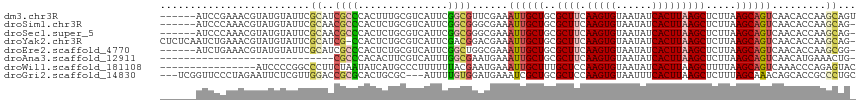
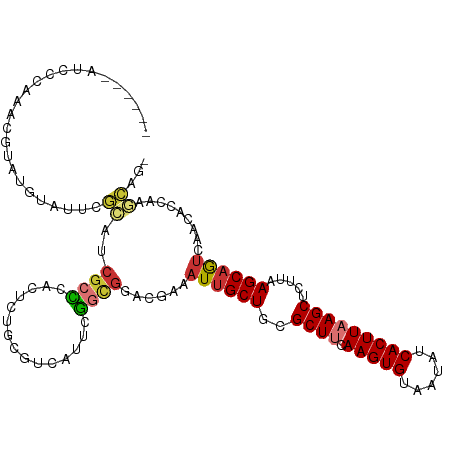

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:10 2011