| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,691,892 – 14,692,022 |
| Length | 130 |
| Max. P | 0.952099 |

| Location | 14,691,892 – 14,692,022 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 151 |
| Reading direction | reverse |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.25952 |
| G+C content | 0.41793 |
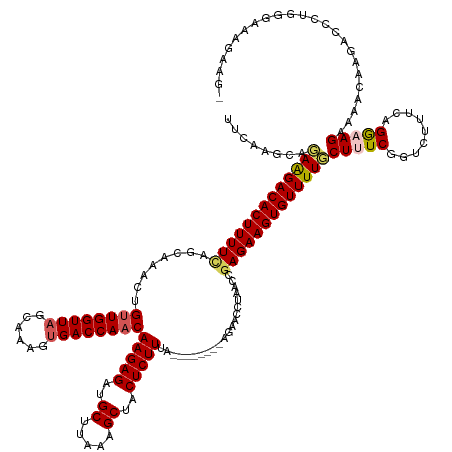
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -30.38 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
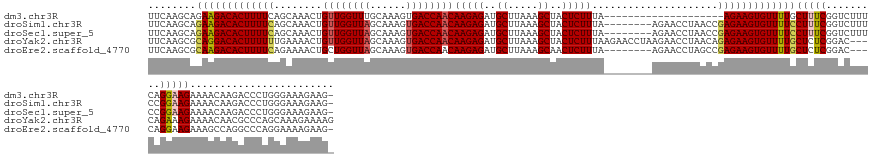
>dm3.chr3R 14691892 130 - 27905053 UUCAAGCAGAAGACACUUUUCAGCAAACUGUUGGUUUGCAAAGUGACCAACAAGAGAUGCUUAAAGCUACUCUUUA--------------------AGAAGUGUUUUGCUUUCGGUCUUUCAGGAAGAAAACAAGACCCUGGGAAAGAAG- ((((((((((..((((((((.........(((((((........)))))))(((((..((.....))..)))))..--------------------)))))))))))))))......(..((((.............))))..)..))).- ( -34.62, z-score = -0.77, R) >droSim1.chr3R 20733615 142 + 27517382 UUCAAGCAGAAGACACUUUUCAGCAAACUGUUGGUUAGCAAAGUGACCAACAAGAGAUGCUUAAAGCUACUCUUUA--------AGAACCUAACCGAGAAGUGUUUUCCUUUCGGUCUUUCCGGAAGAAAACAAGACCCUGGGAAAGAAG- ........(((((((((((((.......(((((((((......)))))))))..((...(((((((.....)))))--------))...))....)))))))))))))(((((((((((..(....).....))))))....)))))...- ( -42.80, z-score = -2.79, R) >droSec1.super_5 662481 142 + 5866729 UUCAAGCAGAAGACACUUUUCAGCAAACUGUUGGUUAGCAAAGUGACCAACAAGAGAUGCUUAAAGCUACUCUUUA--------AGAACCUAACCGAGAAGUGUUUUCCUUUCGGUCUUUCCGGAAGAAAACAAGACCCUGGGAAAGAAG- ........(((((((((((((.......(((((((((......)))))))))..((...(((((((.....)))))--------))...))....)))))))))))))(((((((((((..(....).....))))))....)))))...- ( -42.80, z-score = -2.79, R) >droYak2.chr3R 3355195 148 - 28832112 UUCAAGCGCAGGACACUUUUUUGAAAACUGUUGGUUAGCAAAGUGACCAACAAGAGAUGCUUAAAGCUACUCUUUAAGAACCUAAGAACCUAACAGAGAAGUGUUUUGCUCUCGGAC---CAGAAAGAAAACAACGCCCAGCAAAGAAAAG (((.((.((((((((((((((((.....(((((((((......)))))))))..((...(((((((.....)))))))...))..........)))))))))))))))).)).))).---............................... ( -39.30, z-score = -2.67, R) >droEre2.scaffold_4770 10803890 139 + 17746568 UUCAAGCGCAAGACACUUUUCAGAAAACUGCUGGUUAGCAAAGUGACCAACAAGAGAUGCUUAAAGCAACUCUUUA--------AGAACCUAGCCGAGAAGUGUUUUGCUCUCGGAC---CAGGAAGAAAGCCAGGCCCAGGAAAAGAAG- (((....((((((((((((((((....))...((((((....((.....))(((((.(((.....))).)))))..--------.....))))))))))))))))))))((..((.(---(.((.......)).)).))..))...))).- ( -43.10, z-score = -2.72, R) >consensus UUCAAGCAGAAGACACUUUUCAGCAAACUGUUGGUUAGCAAAGUGACCAACAAGAGAUGCUUAAAGCUACUCUUUA________AGAACCUAACCGAGAAGUGUUUUGCUUUCGGUCUUUCAGGAAGAAAACAAGACCCUGGGAAAGAAG_ ........(((((((((((((........((((((((......))))))))(((((..((.....))..))))).....................)))))))))))))(((((.........)))))........................ (-30.38 = -30.82 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:08 2011