| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,690,907 – 14,691,006 |
| Length | 99 |
| Max. P | 0.607007 |

| Location | 14,690,907 – 14,691,006 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.30 |
| Shannon entropy | 0.54191 |
| G+C content | 0.43197 |
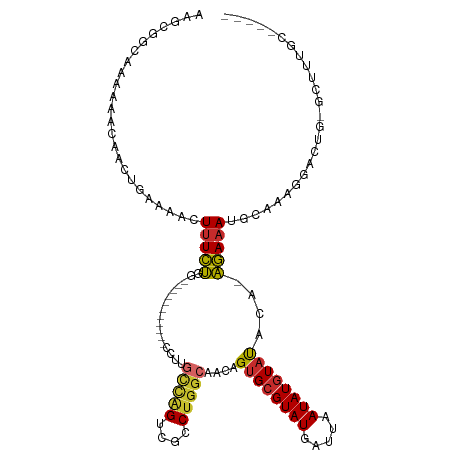
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -12.11 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.21 |
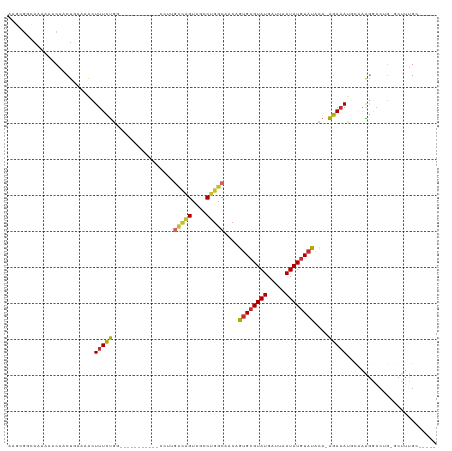
| Combinations/Pair | 1.33 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
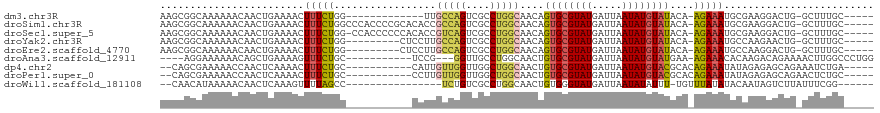
>dm3.chr3R 14690907 99 - 27905053 AAGCGGCAAAAAACAACUGAAAACUUUCUGG-------------UUGCCAGUCGCCUGGCAACAGUGCGUAUGAUUAAUAUGUAUACA-AGAAAUGCGAAGGACUG-GCUUUGC----- .....(((((....(((((((....))).))-------------))((((((((((((....))).))(((((.........))))).-............)))))-)))))))----- ( -28.10, z-score = -1.79, R) >droSim1.chr3R 20732592 112 + 27517382 AAGCGGCAAAAAACAACUGAAAACUUUCUGGCCCACCCCGCACACCGCCAGUCGCCUGGCAACAGUGCGUAUGAUUAAUAUGUAUACA-AGAAAUGCGAAGGACUG-GCUUUGC----- ..((((............(((....))).((....)))))).....((((((((((((....))).))(((((.........))))).-............)))))-)).....----- ( -29.90, z-score = -0.91, R) >droSec1.super_5 661464 111 + 5866729 AAGCGGCAAAAAACAACUGAAAACUUUCUGG-CCACCCCCCACACCGUCAGUCGCCUGGCAACAGUGCGUAUGAUUAAUAUGUAUACA-AGAAAUGCGAAGGACUG-GCUUUGC----- (((((((...........(((....)))..)-))............(((..(((((((....)))(((((((.....)))))))....-......))))..)))..-))))...----- ( -25.72, z-score = -0.10, R) >droYak2.chr3R 3354223 103 - 28832112 AAGCGGCAAAAAACAACUGAAAACUUUCUGG---------CUCCUUGCCAGUCGCCUGGCAACAGUGCGUAUGAUUAAUAUGUAUACA-AGAAAUGCCAAGAACUG-GCUUUGC----- ..(((((...........(((....)))(((---------(.....)))))))))(((....)))(((((((.....)))))))....-......((((.....))-)).....----- ( -27.00, z-score = -1.29, R) >droEre2.scaffold_4770 10802931 103 + 17746568 AAGCGGCAAAAAACAACUGAAAACUUUCUGG---------CUCCUUGCCAGUCGCCUGGCAACAGUGCGUAUGAUUAAUAUGUAUACA-AGAAAUGCCAAGGACUG-GCUUUGC----- .....(((((......(.(((....))).).---------......(((((((.(.(((((...((((((((.....))))))))...-.....))))).))))))-)))))))----- ( -27.90, z-score = -1.18, R) >droAna3.scaffold_12911 3053821 100 - 5364042 ----AGGAAAAAACAGCUGAAAAGUUUCUGC-----------UCCG---GGUUGCCUGGCAACUGUGCGUAUGAUUAAUAUGUAUGAA-AGAAACACAAGACAGAAAACUUGGCCCUGG ----(((................((((((..-----------((..---((((((...))))))((((((((.....)))))))))).-)))))).((((........))))..))).. ( -23.60, z-score = -0.89, R) >dp4.chr2 14815697 101 - 30794189 --CAGCGAAAAACCAACUCAAAACUUUCUGC-----------CAUUGUUGGUUGGCUGGCAACUGUGCGUAUGAUUAAUAUGUACGCACAGAAAUAUAGAGAGCAGAAAUCUGA----- --......................(((((((-----------(.((((..(....)..))))((((((((((.........)))))))))).........).))))))).....----- ( -29.80, z-score = -2.97, R) >droPer1.super_0 6226710 101 + 11822988 --CAGCGAAAAACCAACUCAAAACUUUCUGC-----------CCUUGUUGGUUGGCUGGCAACUGUGCGUAUGAUUAAUAUGUACGCACAGAAAUAUAGAGAGCAGAACUCUGC----- --((((....(((((((..............-----------....))))))).))))....((((((((((.........))))))))))...........((((....))))----- ( -31.87, z-score = -3.34, R) >droWil1.scaffold_181108 2935877 94 - 4707319 --CAACAUAAAAACAACUCAAAGUUUUAGCC----------------UCUGUCGCCUGGCAACUGUGGGUAUGAUUAAUAUAUUU-UGUUUAUAUACAAUAGUCUUAUUUCGG------ --.............(((((.((((...(((----------------..........))))))).)))))..(((((.(((((..-.....)))))...))))).........------ ( -12.90, z-score = 0.29, R) >consensus AAGCGGCAAAAAACAACUGAAAACUUUCUGG___________CCUUGCCAGUCGCCUGGCAACAGUGCGUAUGAUUAAUAUGUAUACA_AGAAAUGCAAAGGACUG_GCUUUGC_____ ........................(((((.................(((((....)))))....((((((((.....))))))))....)))))......................... (-12.11 = -11.90 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:07 2011