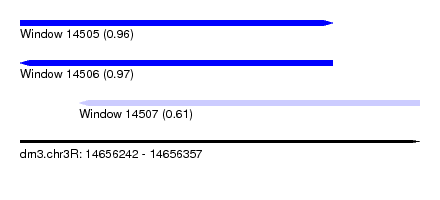
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,656,242 – 14,656,357 |
| Length | 115 |
| Max. P | 0.967166 |

| Location | 14,656,242 – 14,656,332 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.57615 |
| G+C content | 0.50655 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
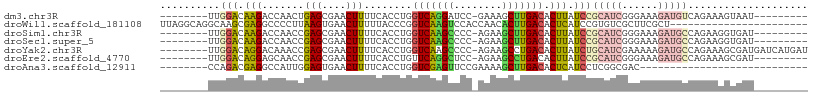
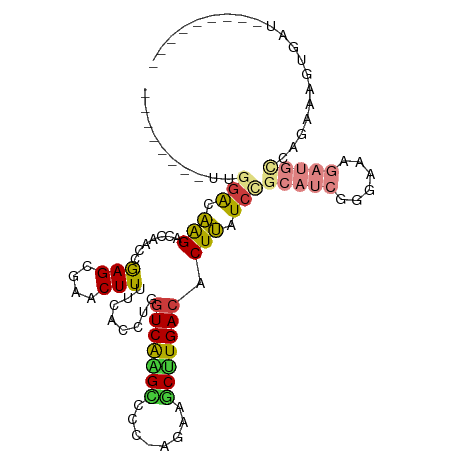
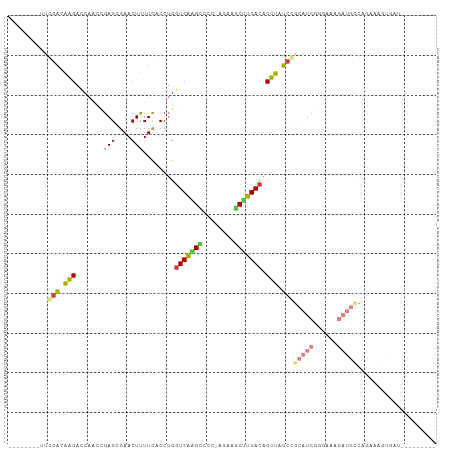
>dm3.chr3R 14656242 90 + 27905053 --------UUGGACAAGACCAACUGAGCGAACUUUUCACCUGGUCAGGAUCC-GAAAGCUUGACACUUAUCCGCAUCGGGAAAGAUGUCAGAAAGUAAU--------- --------((((((..(((((..((((.......))))..)))))..).)))-))....(((((((((.((((...)))).))).))))))........--------- ( -24.90, z-score = -1.79, R) >droWil1.scaffold_181108 2891454 85 + 4707319 UUAGGCAGGCAAGCGAGGCCCCUUAAGUGAACUUUUUACCCGGUCAAGUCCACCAACACUUGACACUCAUCCGUGUCGCUUCGCU----------------------- .......(((((((((.((.....(((....)))........(((((((........)))))))........)).)))))).)))----------------------- ( -22.90, z-score = -1.33, R) >droSim1.chr3R 20696991 90 - 27517382 --------UUGGACAAGACCAACCGAGCGAACUUUUCACCUGGUCAAGCCCC-AGAAGCUUGACACUUAUCCGCAUCGGGAAAGAUGCCAGAAGGUGAU--------- --------((((......)))).............((((((.(((((((...-....)))))))........(((((......)))))....)))))).--------- ( -27.10, z-score = -2.28, R) >droSec1.super_5 623137 90 - 5866729 --------UUGGACAAGACCAACCGAGCGAACUUUUCACCUGGUCAAGCCCC-AGAAGCUUGACACUUAUCCGCAUCGGGAAAGAUGCCAGAAGGUGAU--------- --------((((......)))).............((((((.(((((((...-....)))))))........(((((......)))))....)))))).--------- ( -27.10, z-score = -2.28, R) >droYak2.chr3R 3312903 99 + 28832112 --------UUGGACAGGACAAACCGAGCGAACUUUUCACCUGGUCAAGCCCC-AGAAGCCUGACACUUAUCUGCAUCGAAAAAGAUGCCAGAAAGCGAUGAUCAUGAU --------((((..........)))).........(((..((((((.((..(-((....))).......((((((((......)))).))))..))..))))))))). ( -20.10, z-score = -0.28, R) >droEre2.scaffold_4770 10768095 90 - 17746568 --------UUGGACAGGAGCAACCGAGCGAACUUUUCACCUGUUCAGGCUCC-AGAAGCCUGACACUUAUCCGCAUCGGGAAAGAUGCCAGAAAGCGAU--------- --------..(((((((.((......))((.....)).)))).(((((((..-...)))))))......)))(((((......)))))...........--------- ( -26.30, z-score = -1.38, R) >droAna3.scaffold_12911 3009301 72 + 5364042 --------CCAGACGAGGCCAUUGGAGUGAACUUUUCACCUGGUCGAGUUCCGAAAAGCUUGACACUCAUCCUCGGCGAC---------------------------- --------.....(((((.....((.((((.....)))))).((((((((......))))))))......))))).....---------------------------- ( -20.30, z-score = -0.59, R) >consensus ________UUGGACAAGACCAACCGAGCGAACUUUUCACCUGGUCAAGCCCC_AGAAGCUUGACACUUAUCCGCAUCGGGAAAGAUGCCAGAAAGUGAU_________ ..........(((.(((.......(((.......))).....(((((((........))))))).))).)))(((((......))))).................... (-13.82 = -14.31 + 0.50)
| Location | 14,656,242 – 14,656,332 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.57615 |
| G+C content | 0.50655 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.39 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14656242 90 - 27905053 ---------AUUACUUUCUGACAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUUC-GGAUCCUGACCAGGUGAAAAGUUCGCUCAGUUGGUCUUGUCCAA-------- ---------..(((((((((.((((......)))))))).)))))..........-((((...((((((.(((.........))).))))))..))))..-------- ( -27.10, z-score = -2.21, R) >droWil1.scaffold_181108 2891454 85 - 4707319 -----------------------AGCGAAGCGACACGGAUGAGUGUCAAGUGUUGGUGGACUUGACCGGGUAAAAAGUUCACUUAAGGGGCCUCGCUUGCCUGCCUAA -----------------------(((((.(((((((......))))).......(((((((((...........)))))))))......)).)))))........... ( -26.00, z-score = -0.24, R) >droSim1.chr3R 20696991 90 + 27517382 ---------AUCACCUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUCU-GGGGCUUGACCAGGUGAAAAGUUCGCUCGGUUGGUCUUGUCCAA-------- ---------...........(((((......)))))(((((((..(((((((...-..)))))))((..((((.....))))..)).....)))))))..-------- ( -30.60, z-score = -1.84, R) >droSec1.super_5 623137 90 + 5866729 ---------AUCACCUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUCU-GGGGCUUGACCAGGUGAAAAGUUCGCUCGGUUGGUCUUGUCCAA-------- ---------...........(((((......)))))(((((((..(((((((...-..)))))))((..((((.....))))..)).....)))))))..-------- ( -30.60, z-score = -1.84, R) >droYak2.chr3R 3312903 99 - 28832112 AUCAUGAUCAUCGCUUUCUGGCAUCUUUUUCGAUGCAGAUAAGUGUCAGGCUUCU-GGGGCUUGACCAGGUGAAAAGUUCGCUCGGUUUGUCCUGUCCAA-------- ....(((....(((((((((.((((......)))))))).))))))))(((....-(((((..((((..((((.....))))..)))).))))))))...-------- ( -30.00, z-score = -1.33, R) >droEre2.scaffold_4770 10768095 90 + 17746568 ---------AUCGCUUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAGGCUUCU-GGAGCCUGAACAGGUGAAAAGUUCGCUCGGUUGCUCCUGUCCAA-------- ---------..(((((((((.((((......)))))))).)))))((((((((..-.))))))))(((((.((.....))((......)).)))))....-------- ( -28.80, z-score = -1.12, R) >droAna3.scaffold_12911 3009301 72 - 5364042 ----------------------------GUCGCCGAGGAUGAGUGUCAAGCUUUUCGGAACUCGACCAGGUGAAAAGUUCACUCCAAUGGCCUCGUCUGG-------- ----------------------------...(.(((((..(((((...(((((((((...((.....)).))))))))))))))......))))).)...-------- ( -19.20, z-score = 0.16, R) >consensus _________AUCACUUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUCU_GGGGCUUGACCAGGUGAAAAGUUCGCUCGGUUGGUCUUGUCCAA________ ....................................(((((((.(((((((((....)))))))))..(((((.....)))))........))))))).......... (-18.32 = -18.39 + 0.07)

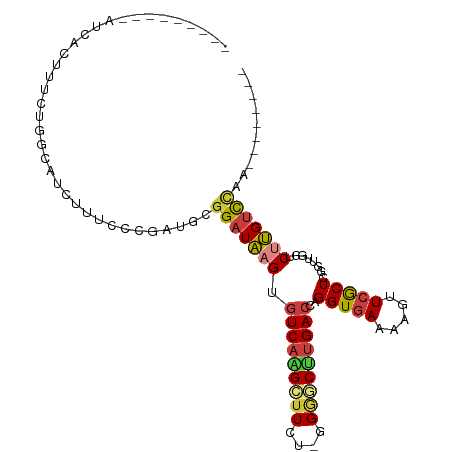
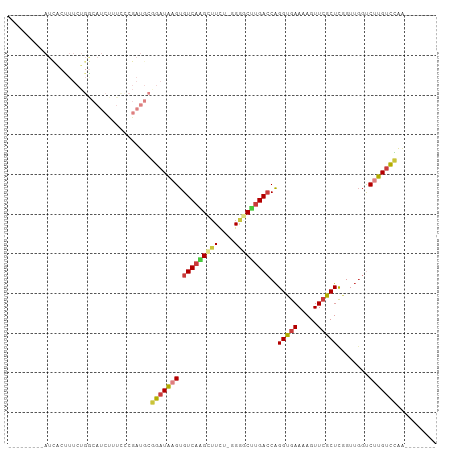
| Location | 14,656,259 – 14,656,357 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.93 |
| Shannon entropy | 0.19175 |
| G+C content | 0.49767 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14656259 98 - 27905053 CCCUUUCCCCAUCAACGUCAUCAUCAUUACUUUCUGACAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUUCGGAUCCUGACCAGGUGAAAAGUUCGCU .........((((...((((..(((.........(((((.(((..(((...)))..))))))))........)))..))))..))))........... ( -17.83, z-score = 0.18, R) >droSim1.chr3R 20697008 98 + 27517382 CCCUUUCACCAUCUUCGUCAUCAUCAUCACCUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUCUGGGGCUUGACCAGGUGAAAAGUUCGCU ...(((((((......................((((.((((......))))))))...(.((((((((.....))))))))).)))))))........ ( -28.50, z-score = -1.93, R) >droSec1.super_5 623154 98 + 5866729 CCCUUUCACCAUCUUCGUCAUCAUCAUCACCUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUCUGGGGCUUGACCAGGUGAAAAGUUCGCU ...(((((((......................((((.((((......))))))))...(.((((((((.....))))))))).)))))))........ ( -28.50, z-score = -1.93, R) >droEre2.scaffold_4770 10768112 94 + 17746568 ----CACCCCAUCAUCCCCAUCAUCAUCGCUUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAGGCUUCUGGAGCCUGAACAGGUGAAAAGUUCGCU ----................(((((..(((((((((.((((......)))))))).)))))((((((((...))))))))...))))).......... ( -28.10, z-score = -1.59, R) >consensus CCCUUUCACCAUCAUCGUCAUCAUCAUCACCUUCUGGCAUCUUUCCCGAUGCGGAUAAGUGUCAAGCUUCUGGAGCCUGACCAGGUGAAAAGUUCGCU ..........................((((((((((.((((......))))))))...(.((((((((.....))))))))))))))).......... (-22.42 = -22.05 + -0.37)

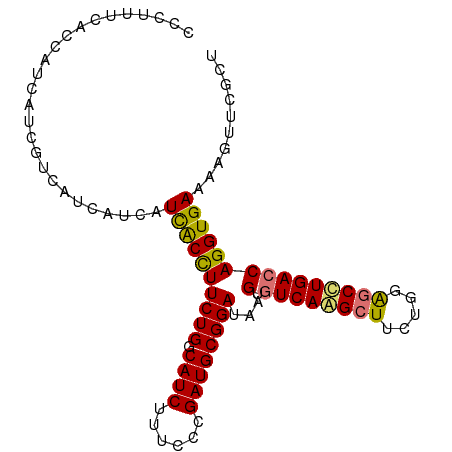
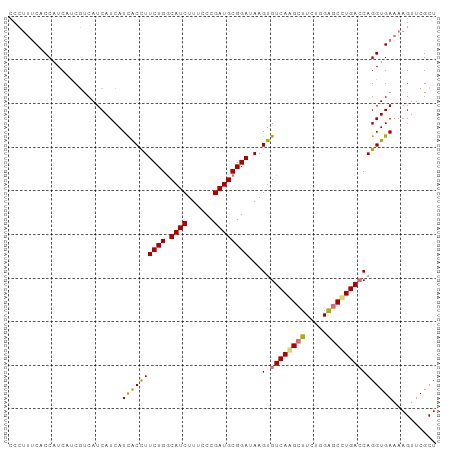
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:04 2011