
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,632,736 – 14,632,802 |
| Length | 66 |
| Max. P | 0.680261 |

| Location | 14,632,736 – 14,632,802 |
|---|---|
| Length | 66 |
| Sequences | 11 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 66.56 |
| Shannon entropy | 0.68859 |
| G+C content | 0.58578 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -9.24 |
| Energy contribution | -8.28 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
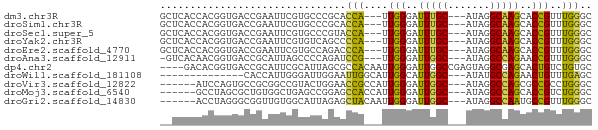
>dm3.chr3R 14632736 66 + 27905053 GCUCACCACGGUGACCGAAUUCGUGCCCGCACCA---UUGGGAUUUGC---AUAGGCAAGCACCGUUUGGGC (((((..((((((.((.(((..(((....))).)---)).)).(((((---....))))))))))).))))) ( -23.70, z-score = -1.39, R) >droSim1.chr3R 20674108 66 - 27517382 GCUCACCACGGUGACCGAAUUCGUGCCCGCACCA---UUGGGAUUUGC---AUAGGCAAGCACCGUUUGGGC (((((..((((((.((.(((..(((....))).)---)).)).(((((---....))))))))))).))))) ( -23.70, z-score = -1.39, R) >droSec1.super_5 599475 66 - 5866729 GCUCACCACGGUGACCGAAUUCGUGCCCGUACCA---UUGGGAUUUGC---AUAGGCAAGCACCGUUUGGGC (((((..((((((............((((.....---.)))).(((((---....))))))))))).))))) ( -22.40, z-score = -1.12, R) >droYak2.chr3R 3291677 66 + 28832112 GCUCACCACGGUGACCGAAUUCGUGUCAGCCCCA---UUGGGAUUUGC---AUAGGCAAGCACCGUUUGGGC (((((..((((((((((....)).))).(((((.---..)))..((((---....))))))))))).))))) ( -22.80, z-score = -1.26, R) >droEre2.scaffold_4770 10744330 66 - 17746568 GCUCACCACGGUGACCGAAUUCGUGCCAGACCCA---UUGGGAUUUGC---AUAGGCAAGCACCGUUUGGGC (((((..((((((.(((....)((((.((((((.---..))).)))))---)).))....)))))).))))) ( -21.40, z-score = -1.00, R) >droAna3.scaffold_12911 2986503 65 + 5364042 -GUCACAACGGUGACCGCAUUAGCCCCAGAUCCG---UUGGGAUUGGC---AUAGGCCAGAACCGUUUGGGC -(((((....))))).((....))(((((((...---..((..(((((---....)))))..))))))))). ( -22.50, z-score = -1.25, R) >dp4.chr2 14759257 68 + 30794189 ----GACACGGUGACCGCAUUCGCAUUAGCGCCACAAUUGGGAUUGGCCGAGUAGGCGAGCACUGUCUGUGC ----...((((((..(((((((((....))((((..........)))).))))..)))..))))))...... ( -19.30, z-score = 0.49, R) >droWil1.scaffold_181108 2864199 55 + 4707319 --------------CACCAUUGGGAUUGGAAUUGGCAUUGGCAUUGGC---AUAUGCCAGAACUGUUUGAGC --------------..(((.......)))..(((((((..((....))---..)))))))............ ( -12.60, z-score = -0.04, R) >droVir3.scaffold_12822 3168513 63 + 4096053 ------AUCCAGUGCCGCGGCCGUACUGGAACCGCCAUUGGGAUUGGC---AUAGGCCAGCGCCGCCUGGGC ------.(((((.(((((((((.....(....)((((.......))))---...)))).)))..))))))). ( -26.10, z-score = -0.02, R) >droMoj3.scaffold_6540 19076121 63 - 34148556 ------GCCUAGCGCUGUGGCUGAGCCGGAGCCACCAUUGGGAUUGGC---AUAGGCCAGCACCGUCUGGGC ------((((((.((.(((.(((.(((...((((((....))..))))---...))))))))).)))))))) ( -29.70, z-score = -1.47, R) >droGri2.scaffold_14830 2581198 63 - 6267026 ------ACCUAGGGCGGUUGUGGCAUUAGAGCUACAAUUGGGAUUGGC---AUAGGCCAAUGCCGUUUGGGC ------.((((((.((((((((((......)))))))))((.((((((---....)))))).))))))))). ( -27.30, z-score = -3.64, R) >consensus __UCACCACGGUGACCGAAUUCGUGCCAGAACCA___UUGGGAUUGGC___AUAGGCAAGCACCGUUUGGGC ...............................(((....(((..(((((.......)))))..)))..))).. ( -9.24 = -8.28 + -0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:26:02 2011