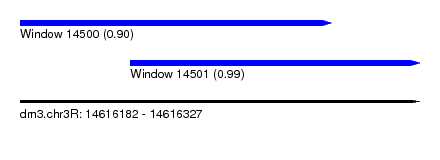
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,616,182 – 14,616,327 |
| Length | 145 |
| Max. P | 0.985634 |

| Location | 14,616,182 – 14,616,295 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Shannon entropy | 0.19210 |
| G+C content | 0.34615 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
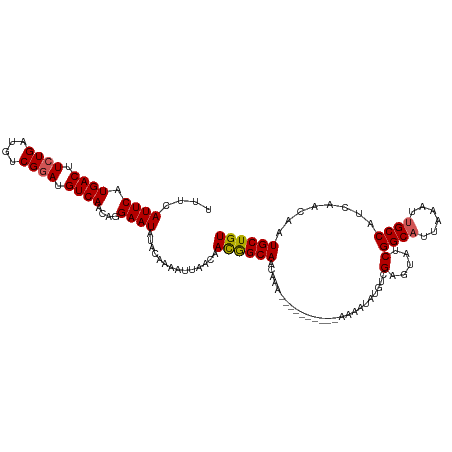
>dm3.chr3R 14616182 113 + 27905053 UUUCAUUCAUGACUUCUGAUGUCGGAUGUCAACAGGAAUAUACAAAAUUAACAACGGCAACAAAAAAA------AAAAAAUGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGU ....((((.((((.((((....)))).))))....))))..............(((((..........------....(((((((.....)))))))..((((.......))))))))) ( -23.30, z-score = -2.06, R) >droSim1.chr3R 20657735 119 - 27517382 UUUCAUUCAUGACUUCUGAUGUCGGAUGUCAACAGGAAUAUACAAAAUUAACAACGGCACCAAAAAAAAAAAAAAAAAUAUGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGU ....((((.((((.((((....)))).))))....))))..............((((((..................((((.....)))).((((......))))........)))))) ( -22.10, z-score = -1.62, R) >droSec1.super_5 583103 109 - 5866729 UUUCAUUCAUGACUUCUGAUGUCGGAUGUCAACAGGAAUAUACAAAAUUAACAAUGGCAACAAA----------AAAAUAUGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGU ....((((.((((.((((....)))).))))....))))..(((..(((....(((((((....----------.....((((((.....))))))....)))))))....)))..))) ( -23.72, z-score = -1.87, R) >droYak2.chr3R 3273774 105 + 28832112 UUUCAUUCAUGACUUCUGAUGUCGCAUGUCAACAGGAAUAUACAAAAUUAACAACAGCAACAAA-----------AAAC---UCGAGUAUCGGCAUUAAAUCGCCAUCAACAAUGCUGU .............(((((.((.(....).)).)))))................((((((.....-----------..((---....))...(((........)))........)))))) ( -15.70, z-score = -0.32, R) >droEre2.scaffold_4770 10728017 104 - 17746568 UUUCAUUCAUGACUUCUGAUGUCGGAUGUCAACAGGAAUAUAUAUAAA-ACGAACAGCAGCAAA-----------AAAG---UCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGU ....((((.((((.((((....)))).))))....)))).........-.......((((((..-----------....---..(.....)((((......))))........)))))) ( -23.00, z-score = -1.60, R) >consensus UUUCAUUCAUGACUUCUGAUGUCGGAUGUCAACAGGAAUAUACAAAAUUAACAACGGCAACAAA__________AAAAUAUGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGU ....((((.((((.((((....)))).))))....))))..............((((((.........................(.....)((((......))))........)))))) (-20.18 = -20.18 + -0.00)

| Location | 14,616,222 – 14,616,327 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.59 |
| Shannon entropy | 0.22212 |
| G+C content | 0.33619 |
| Mean single sequence MFE | -18.89 |
| Consensus MFE | -17.46 |
| Energy contribution | -17.66 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
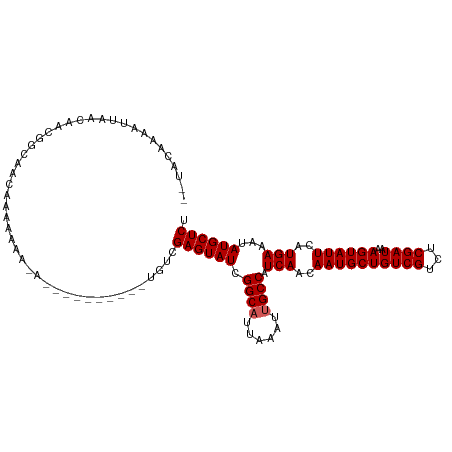
>dm3.chr3R 14616222 105 + 27905053 --UACAAAAUUAACAACGGCAACAAAAAAAAAAAAA------UGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGUCGUCUCGAUAAAGUAUUCAUGAAAUAUGCUCU --...............((((...............------))))((((((.((((......)))).(((..((((((((((...))))..))))))..)))...)))))). ( -19.76, z-score = -2.66, R) >droSim1.chr3R 20657775 111 - 27517382 --UACAAAAUUAACAACGGCACCAAAAAAAAAAAAAAAAAUAUGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGUCGUCUCGAUAAAGUAUUCAUGAAAUAUGCUCU --...............((((.....................))))((((((.((((......)))).(((..((((((((((...))))..))))))..)))...)))))). ( -19.20, z-score = -2.54, R) >droSec1.super_5 583143 101 - 5866729 --UACAAAAUUAACAAUGGCAACAAAAAAAUA----------UGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGUCGUCUCGAUAAAGUAUUCAUGAAAUAUGCUCU --..............((....))........----------....((((((.((((......)))).(((..((((((((((...))))..))))))..)))...)))))). ( -20.30, z-score = -2.46, R) >droYak2.chr3R 3273814 97 + 28832112 --UACAAAAUUAACAACAGCAACAAAAAAC--------------UCGAGUAUCGGCAUUAAAUCGCCAUCAACAAUGCUGUCGUCUCGAUAAAGUAUUCAUGAAAUAUGCUCU --............................--------------..((((((.(((........))).(((..((((((((((...))))..))))))..)))...)))))). ( -16.40, z-score = -2.13, R) >droEre2.scaffold_4770 10728057 96 - 17746568 UAUAUAAAACGAACAGCAGCAAAAAAG-----------------UCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGUCGUCUCGAUCAAGUAUUCAUGAAAUAUGCUCU .........(((((.((((((......-----------------..(.....)((((......))))........)))))).)).)))....(((((.........))))).. ( -18.80, z-score = -1.49, R) >consensus __UACAAAAUUAACAACGGCAACAAAAAAA_A__________UGUCGAGUAUCGGCAUUAAAUUGCCAUCAACAAUGCUGUCGUCUCGAUAAAGUAUUCAUGAAAUAUGCUCU ..............................................((((((.((((......)))).(((..((((((((((...))))..))))))..)))...)))))). (-17.46 = -17.66 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:59 2011