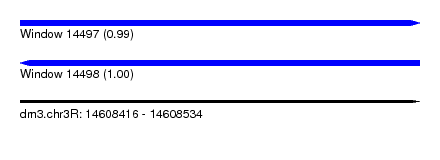
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,608,416 – 14,608,534 |
| Length | 118 |
| Max. P | 0.999525 |

| Location | 14,608,416 – 14,608,534 |
|---|---|
| Length | 118 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 64.42 |
| Shannon entropy | 0.73526 |
| G+C content | 0.34121 |
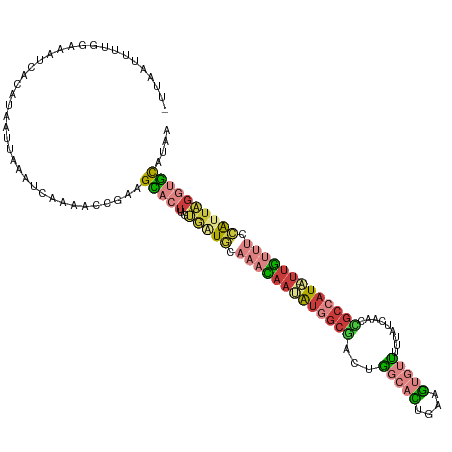
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -9.21 |
| Energy contribution | -10.86 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14608416 118 + 27905053 UUAUGCACCUAAUGGAAACAA-UAUGGCGAUUGAUAAAAACACUUCAGUGCCAGUCGCCAUUUUGUUUGCAACACAAGUGCUUUGGUUUUGAUUUAAUUAUGUGAUUUCCGAAAUUAAA ....((((....((.((((((-.((((((((((.......(((....))).)))))))))).)))))).))......)))).(((((((((....((((....))))..))))))))). ( -31.20, z-score = -2.78, R) >droSim1.chr3R 20650069 118 - 27517382 UUAUGCACCUAAUGGAAACAA-UAUGGCGGUUGAUAAAAACACUUCAGUGCCAGUCGCCAUAUUGUUUGCAUCACAAGUGCUUCGGUUUUGAUUUAAUUAUGUGAUUUCCAAAAUUAAA ....((((...(((.((((((-(((((((..((.......(((....))).))..))))))))))))).))).....))))...(((((((....((((....))))..)))))))... ( -32.50, z-score = -2.97, R) >droSec1.super_5 574384 118 - 5866729 UUAUGCACCUAAUGGAAACAA-UAUGGCAGUUGAUAAAAACACUUCAGUGCCGGUCGCCAUAUUGUUUGCAUCACCAGUGCUUCGGUUUUGAUUUAAUUAUGUGAUUUCCAAAAUUAAA ..((((.((....))((((((-((((((....(((.....(((....)))...)))))))))))))))))))...(((.((....)).))).(((((((.((.(....))).))))))) ( -28.70, z-score = -1.70, R) >droYak2.chr3R 3266219 117 + 28832112 UUCUGCACCUAAUGGAAACAA-UAUGGCGGUUGAUAAAAACACUCCAGUGCCAGUCGCCAUAUUGUUUGCAUCACAAGUGCUUCGGUUUUGAUUUAAUUAUGUGAUUUCGAAAUUAAA- ....((((...(((.((((((-(((((((..((.......(((....))).))..))))))))))))).))).....))))...((((((((.(((......)))..))))))))...- ( -33.90, z-score = -3.30, R) >droEre2.scaffold_4770 10720796 112 - 17746568 UUAUGCACCUAAUGGAAACAA-UAUGGCGGUUGAUAAAAACACUCCAGUGCCCGUCGCCAUAUUGUUUGCAUCACAAGUGCUUUGGUUUUGAUUUAAUUAUGUGAUUUCCAAA------ ....((((...(((.((((((-((((((((((......)))((....))......))))))))))))).))).....)))).((((...(.((........)).)...)))).------ ( -29.70, z-score = -2.03, R) >droAna3.scaffold_12911 2963788 98 + 5364042 UUAUAAAACAAACCGCCAUUA-AACCGCCAUUUUUCAAUGCAAUUCACACAAAAUUGC--UCUUAAUUUCAGAUUAAAGUCUGCCAAAUUGAGA-AAUUAAA----------------- .....................-.................((((((.......))))))--((((((((((((((....)))))..)))))))))-.......----------------- ( -14.60, z-score = -2.51, R) >dp4.chr2 14736911 117 + 30794189 UUCGACUUUUAAUCAUGGCAGUAAUUGUUCUUUUUUGAGAGUACCUAUGUACAUGACUCACAUUAGUCAGACUCUUUUUACUUUAUCAU--AAAUAACCACCUAUCUUUCAGAGAUAUU ...((((.....(((((...(((..(((((((....)))))))..)))...)))))........)))).....................--...........((((((...)))))).. ( -14.82, z-score = 0.46, R) >consensus UUAUGCACCUAAUGGAAACAA_UAUGGCGGUUGAUAAAAACACUUCAGUGCCAGUCGCCAUAUUGUUUGCAUCACAAGUGCUUCGGUUUUGAUUUAAUUAUGUGAUUUCCAAAAUUAA_ ....((((....((.((((((...(((((((((..................)))))))))..)))))).))......))))...................................... ( -9.21 = -10.86 + 1.64)

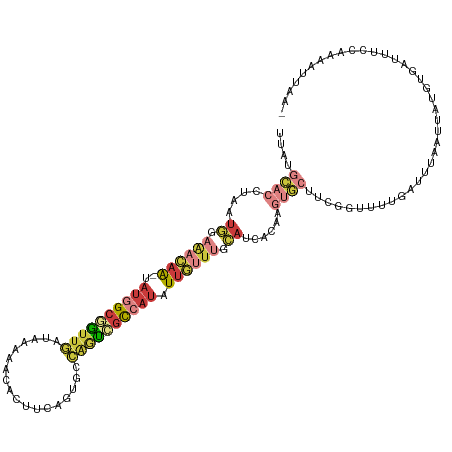
| Location | 14,608,416 – 14,608,534 |
|---|---|
| Length | 118 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.42 |
| Shannon entropy | 0.73526 |
| G+C content | 0.34121 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.94 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14608416 118 - 27905053 UUUAAUUUCGGAAAUCACAUAAUUAAAUCAAAACCAAAGCACUUGUGUUGCAAACAAAAUGGCGACUGGCACUGAAGUGUUUUUAUCAAUCGCCAUA-UUGUUUCCAUUAGGUGCAUAA (((((((..(.......)..)))))))...........(((((((...((.((((((.(((((((.((((((....))))......)).))))))).-)))))).)).))))))).... ( -31.10, z-score = -3.29, R) >droSim1.chr3R 20650069 118 + 27517382 UUUAAUUUUGGAAAUCACAUAAUUAAAUCAAAACCGAAGCACUUGUGAUGCAAACAAUAUGGCGACUGGCACUGAAGUGUUUUUAUCAACCGCCAUA-UUGUUUCCAUUAGGUGCAUAA (((((((.((.......)).)))))))...........(((((..(((((.(((((((((((((..((((((....))))......))..)))))))-)))))).)))))))))).... ( -35.60, z-score = -4.37, R) >droSec1.super_5 574384 118 + 5866729 UUUAAUUUUGGAAAUCACAUAAUUAAAUCAAAACCGAAGCACUGGUGAUGCAAACAAUAUGGCGACCGGCACUGAAGUGUUUUUAUCAACUGCCAUA-UUGUUUCCAUUAGGUGCAUAA (((((((.((.......)).)))))))...........(((((..(((((.((((((((((((((..(((((....)))))....))....))))))-)))))).)))))))))).... ( -32.60, z-score = -3.10, R) >droYak2.chr3R 3266219 117 - 28832112 -UUUAAUUUCGAAAUCACAUAAUUAAAUCAAAACCGAAGCACUUGUGAUGCAAACAAUAUGGCGACUGGCACUGGAGUGUUUUUAUCAACCGCCAUA-UUGUUUCCAUUAGGUGCAGAA -(((((((..(......)..)))))))...........(((((..(((((.(((((((((((((..((((((....))))......))..)))))))-)))))).)))))))))).... ( -32.70, z-score = -3.34, R) >droEre2.scaffold_4770 10720796 112 + 17746568 ------UUUGGAAAUCACAUAAUUAAAUCAAAACCAAAGCACUUGUGAUGCAAACAAUAUGGCGACGGGCACUGGAGUGUUUUUAUCAACCGCCAUA-UUGUUUCCAUUAGGUGCAUAA ------(((((......................)))))(((((..(((((.((((((((((((((..(((((....)))))..)......)))))))-)))))).)))))))))).... ( -33.05, z-score = -3.31, R) >droAna3.scaffold_12911 2963788 98 - 5364042 -----------------UUUAAUU-UCUCAAUUUGGCAGACUUUAAUCUGAAAUUAAGA--GCAAUUUUGUGUGAAUUGCAUUGAAAAAUGGCGGUU-UAAUGGCGGUUUGUUUUAUAA -----------------..(((((-((.......((....)).......)))))))(((--((((..((((.(((((((((((....))).))))))-))...)))).))))))).... ( -18.14, z-score = -0.31, R) >dp4.chr2 14736911 117 - 30794189 AAUAUCUCUGAAAGAUAGGUGGUUAUUU--AUGAUAAAGUAAAAAGAGUCUGACUAAUGUGAGUCAUGUACAUAGGUACUCUCAAAAAAGAACAAUUACUGCCAUGAUUAAAAGUCGAA ..(((((.....)))))((..((..(((--((......)))))..(((..(((((......))))).((((....)))).)))..............))..)).((((.....)))).. ( -19.80, z-score = -0.22, R) >consensus _UUAAUUUUGGAAAUCACAUAAUUAAAUCAAAACCGAAGCACUUGUGAUGCAAACAAUAUGGCGACUGGCACUGAAGUGUUUUUAUCAACCGCCAUA_UUGUUUCCAUUAGGUGCAUAA ......................................(((((..(((((.(((((((((((((...(((((....))))).........))))))).)))))).)))))))))).... (-13.30 = -14.94 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:57 2011