| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,593,823 – 14,593,929 |
| Length | 106 |
| Max. P | 0.928247 |

| Location | 14,593,823 – 14,593,929 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.64 |
| Shannon entropy | 0.70158 |
| G+C content | 0.47708 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.96 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
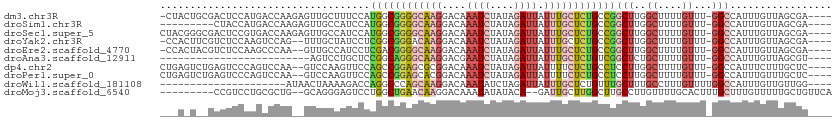
>dm3.chr3R 14593823 106 + 27905053 -CUACUGCGACUCCAUGACCAAGAGUUGCUUUCCAUGGCGGGGCAAGGACAAAUCUAUAGAUUAUUUGCUCUGCCGGCUUGGCUUUUGUUU-GGCCAUUUGUUAGCGA---- -.....(((((((.........))))))).......(((((((((((....((((....)))).)))))))))))((((..((....))..-))))............---- ( -36.80, z-score = -2.41, R) >droSim1.chr3R 20626857 98 - 27517382 ---------CUACCAUGACCAAGAGUUGCCAUCCAUGGCGGGGCAAGGACAAAUCUAUAGAUUAUUUGCUCUGCCGGCUUGGCUUUUGUUU-GGCCAUUUGUUAGCGA---- ---------.....(((.(((((....((((....((((((((((((....((((....)))).))))))))))))...)))).....)))-)).)))..........---- ( -33.40, z-score = -1.88, R) >droSec1.super_5 560078 107 - 5866729 CUACGGGCGACUCCGUGACCAAGAGUUGCCAUCCAUGGCGGGGCAAGGACAAAUCUAUAGAUUAUUUGCUCUGCCGGCUUGGCUUUUGUUU-GGCCAUUUGUUAGCGA---- ...((((((((((.........))))))))......(((((((((((....((((....)))).)))))))))))((((..((....))..-)))).........)).---- ( -41.10, z-score = -2.68, R) >droYak2.chr3R 3252656 104 + 28832112 -CCACUUCGUCUCCAAGUCCAG--UUUGCUAUCCUCGGCGGGACAAGGACAAAUCUAUAGAUUAUUUGCUCUGCCGGCUUGGCUUUUGUUU-GGCCAUUUGUUAGCGA---- -.....((((...(((((((((--...((((..(.((((((..((((....((((....)))).))))..)))))))..))))......))-))..)))))...))))---- ( -28.60, z-score = -0.74, R) >droEre2.scaffold_4770 10706180 104 - 17746568 -CCACUACGUCUCCAAGCCCAA--GUUGCCAUCCUCGACGGGGCAAGGACAAAUCUAUAGAUUAUUUGCUCUGCCGGCUUGGCUUUUGUUU-GGCCAUUUGUUAGCGA---- -......(((...(((((((((--(..((((..(.((.(((((((((....((((....)))).))))))))).)))..)))).))))...-)))...)))...))).---- ( -28.70, z-score = -0.36, R) >droAna3.scaffold_12911 2949780 82 + 5364042 -------------------------AGUCCUGCUCCGGCAGGGCAAGGACGAAUCUAUAGAUUAUUUGCUCUGUCGGCUCUGCUUUUGUUU-GGCCAUUUGUUAGCGU---- -------------------------......(((..(((((((((((....((((....)))).)))))))))))((((..((....))..-)))).......)))..---- ( -29.30, z-score = -2.44, R) >dp4.chr2 14721656 105 + 30794189 CUGAGUCUGAGUCCCAGUCCAA--GUCCAAGUUCCAGCCGGAGCGCGGACAAAUCUAUAGAUUAUUUUCUCUGCCUCCUUGGCUUUUGUUU-GGCCAUUUCUUUGCUC---- ..((((..(((.....(.((((--((((..(((((....)))))..)))).((((....)))).........(((.....)))......))-))).....))).))))---- ( -26.40, z-score = -0.81, R) >droPer1.super_0 6130541 105 - 11822988 CUGAGUCUGAGUCCCAGUCCAA--GUCCAAGUUCCAGCCGGAGCACGGACAAAUCUAUAGAUUAUUUUCUCUGCCUCCUUGGCUUUUGUUU-GGCCAUUUGUUUGCUC---- ..(((.(.(((...........--((((..(((((....)))))..)))).((((....)))).....))).).)))..(((((.......-)))))...........---- ( -25.10, z-score = -0.49, R) >droWil1.scaffold_181108 2821641 87 + 4707319 ---------------------AUAACUAAAAGACCAGGCCCAGCAAGGACAAAUAUCUAGAUUAUUUGCUCUGUUUGCUUUGCCUUUGUUUUGGCCAUUUGUUGUUGG---- ---------------------....((((((.(..((((..((((((((((((((.......)))))).)))..)))))..)))).).))))))(((........)))---- ( -20.30, z-score = -0.70, R) >droMoj3.scaffold_6540 19030115 99 - 34148556 ---------CCGUCCUGCGCUG--GCAGGGAGUCCUGGCUGAACAAGGACAAAUAUAUACA--GAUUGCUUGGCUUGCCUUGUUUUGCACUUUGCUUUGUUUUUGCUGUUCA ---------.(..(((((....--)))))..)....(((.((((((((.((((......((--((..((..((....))..))))))...))))))))))))..)))..... ( -25.20, z-score = 0.63, R) >consensus ___A_U__GACUCCAAGACCAA__GUUGCCAUCCCUGGCGGGGCAAGGACAAAUCUAUAGAUUAUUUGCUCUGCCGGCUUGGCUUUUGUUU_GGCCAUUUGUUAGCGA____ ...................................((((((((((((....((((....)))).)))))))))))).....(((........)))................. (-13.68 = -13.96 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:55 2011