| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,582,324 – 14,582,441 |
| Length | 117 |
| Max. P | 0.610257 |

| Location | 14,582,324 – 14,582,441 |
|---|---|
| Length | 117 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.35 |
| Shannon entropy | 0.71704 |
| G+C content | 0.53600 |
| Mean single sequence MFE | -42.19 |
| Consensus MFE | -10.96 |
| Energy contribution | -10.66 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.94 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
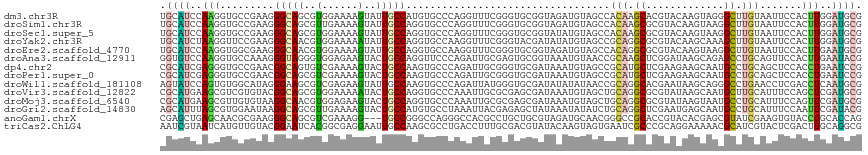
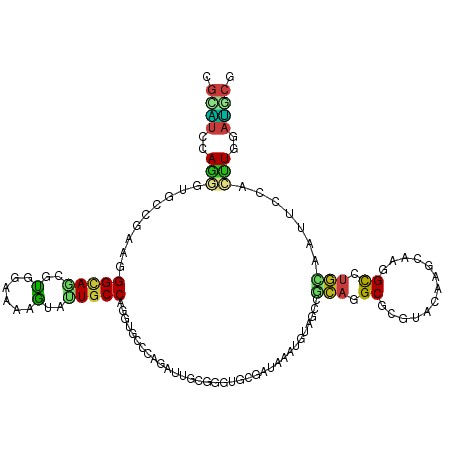
>dm3.chr3R 14582324 117 + 27905053 UGCAUCCAAGGUGCCGAAGGGCAGCGUGGAAAAGUAUUGCCAUGUGCCCAGGUUUCGGGUGCGGUAGAUGUAGCCACAAGCACGUACAAGUAGGGCUUGUAAUUCCACUUGGAUGCG .(((((((((.((((....)))).)((((((.....(((((..(..(((.......)))..))))))........((((((.(.((....)).)))))))..)))))))))))))). ( -52.60, z-score = -4.33, R) >droSim1.chr3R 20615571 117 - 27517382 UGCAUCCAAGGUGCCGAAGGGCAGCGUUGAAAAGUAUUGCCAGGUGCCCAGGUUUCGGGUGCGGUAGAUGUAGCCACAAGCGCGUACAAGUAAGGCUUGUAAUUCCACUUGGAUGCG .(((((((((.((((....))))(((((........(((((..(..(((.......)))..)))))).(((....)))))))).((((((.....))))))......))))))))). ( -48.00, z-score = -3.21, R) >droSec1.super_5 548800 117 - 5866729 UGCAUCCAAGGUGCCGAAGGGCAGCGUGGAAAAGUAUUGCCAGGUGCCCAGGUUUCGGGUGCGGUAUAUGUAGCCACAAGCGCGUACAAGUAAGGCUUGUAAUUCCACUUGGAUGCG .(((((((((.((((....)))).)((((((.......(((..(..(((.......)))..)(((.......)))....).)).((((((.....)))))).)))))))))))))). ( -50.20, z-score = -3.64, R) >droYak2.chr3R 3240411 117 + 28832112 UGCAUCUAAGGUUCCGAAGGGCAACGUGGAAAAAUAUUGCCAGGUGCCAAGGUUUCGGGUACGAUAUAUGUAGCCGCAGGCGCGUACAAGCAAAGCUUGUAAUUCCACUUGGAUGCG .((((((((((((((....)).))).(((((............(((((..(((((((....))).......))))...))))).(((((((...))))))).)))))))))))))). ( -44.41, z-score = -3.26, R) >droEre2.scaffold_4770 10694916 117 - 17746568 UGCAUCCAAGGUGGCGAAGGGCAACGUGGAAAAGUAUUGCCAGGUGCCAAGGUUUCGGGUGCGGUAGAUGUAGCCACAGGCGCGUACAAGUAAGGCUUGUAAUUCCACUUGAAUGCG .((((.(((((((((....(((((..(......)..)))))..(..((.........))..)..........))))).((....((((((.....))))))...)).)))).)))). ( -39.20, z-score = -0.98, R) >droAna3.scaffold_12911 2938900 117 + 5364042 GGUGUCCAAGGUGCCAAAGGGUAGGGUGGAGAAGUACUGCCAGGUUCCCAGAUUGCGAGUGCGGUAAAUGUAACCGCAAGCUCGGAUAAGCAGAGCCUGCAGUUCCACUUGAAUACG .((((.((((.((((....))))...((((.....(((((.((((((......(.((((((((((.......)))))..))))).)......))))))))))))))))))).)))). ( -42.90, z-score = -2.30, R) >dp4.chr2 16521576 117 + 30794189 CGCAUCGAGGGUGCCGAACGGCAGUGUCGAAAAGUACUGCCAAGUGCCCAGAUUGCGGGUGCGAUAAAUGUAGCCGCAUGCUCGAAGAAGCAAUGCCUGCAGCUCCACCUGAAUCCG ........(((..(.....(((((((.(.....))))))))..)..))).((((.((((((.((....(((((..(((((((......))).)))))))))..)))))))))))).. ( -42.90, z-score = -1.96, R) >droPer1.super_0 7947917 117 - 11822988 CGCAUCGAGGGUGCCGAACGGCAGCGUCGAAAAGUACUGCCAAGUGCCCAGAUUGCGGGUGCGAUAAAUGUAGCCGCAUGCUCGAAGAAGCAAUGCCUGCAGCUCCACCUGAAUCCG ....((((.(.((((....)))).).))))...(((((....)))))...((((.((((((.((....(((((..(((((((......))).)))))))))..)))))))))))).. ( -43.40, z-score = -1.82, R) >droWil1.scaffold_181108 2809768 117 + 4707319 AGUAUCCAGUGUGGCAUAGGGAAGCGUCGAGAAGUAUUGCCAAGUGCCCAGAUUAUGGGUGCGAUAUAUAUAACCGCAGGCACGAAUAAGCAGGGCCUGAACCUCGACCUCAAUGCG .............((((.(((....((((((..(((((((.....(((((.....)))))))))))).........(((((.(.........).)))))...))))))))).)))). ( -35.70, z-score = -0.96, R) >droVir3.scaffold_12822 3109546 117 + 4096053 CGCAUGAAGCGUCGUGUACGGCAGCGUGGAAAAAUACUGCCAGGUGCCCAAAUUGCGCGAGCGAUAAAUGUAGCUGCAGGCGCGUAUAAGCAAUGCUUGCAUUUCCAGCUCGAUGCG (((((..(((((((....))))....((((((....((((.((.(((....(((((....)))))....))).))))))(((.((((.....)))).))).)))))))))..))))) ( -43.40, z-score = -0.80, R) >droMoj3.scaffold_6540 19016380 117 - 34148556 CGCAUGAAGCGUUGUGUAAGGCAACGUGGAGAAGUACUGCCAGGUGCCCAAAUUGCGCGAGCGAUAAAUGUAGCUGCAGGCGCGUAUAAGUAAUGCCUGCAUUUCCAGUUCGAUGCG (((((((((((((((.....)))))))(((((.(((((....)))))....(((((....))))).........(((((((((......))...))))))))))))..))).))))) ( -42.20, z-score = -1.64, R) >droGri2.scaffold_14830 2533166 117 - 6267026 AGCAUUUAGCGUGGAAUAAGGCAGCGUUGAAAAGUACUGCCAUGUGCCUAAAUUACGAGAGCUAUAAAUAUAUCUGCAGGCUCGAAUGAGCAAUGCCUGCAUUUCCAAUUCGAUACG .((.....)).(((((...(((((..((....))..))))).(((((((........)).)).)))........(((((((.............))))))).))))).......... ( -30.02, z-score = -0.66, R) >anoGam1.chrX 21852470 114 - 22145176 CGAGCUGAGCAACGCGAAGGGCAGCGUCGAAAGG---UGCCGGGCCAGGGCCACGCCUGCUGCGUAGAUGCAACGGGCCGCACCGUACACGAGCGUAUCGAAGUGUACCUGCACCAG ...((........))...((((((........((---(((..(((....)))..(((((.((((....)))).))))).)))))((((((...((...))..)))))))))).)).. ( -44.10, z-score = 0.31, R) >triCas2.ChLG4 11289619 117 - 13894384 AAUCGUAAUCAUGUUGUACGGAAUCACGGCGAGGAAUUGCCAAGCGCCUGACCUUUGCGACGUAUACAAGUAGUGAAUCGCCCGCAGGAAAAACGCAUCGUACUCGACUUGCAGGCG ....(((...((((((((.((..(((.((((.((.....))...))))))).)).)))))))).)))...........((((.(((((.........(((....)))))))).)))) ( -31.60, z-score = 0.26, R) >consensus CGCAUCCAGGGUGCCGAAGGGCAGCGUGGAAAAGUAUUGCCAGGUGCCCAGAUUGCGGGUGCGAUAAAUGUAGCCGCAGGCGCGUACAAGCAAGGCCUGCAAUUCCACUUGGAUGCG .((((..(((.........(((((..(......)..)))))..................................(((.((.............)).))).......)))..)))). (-10.96 = -10.66 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:53 2011