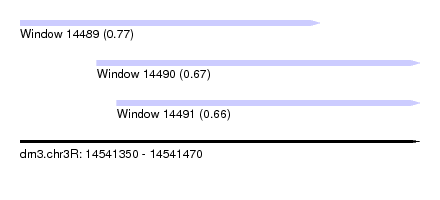
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,541,350 – 14,541,470 |
| Length | 120 |
| Max. P | 0.770609 |

| Location | 14,541,350 – 14,541,440 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Shannon entropy | 0.44552 |
| G+C content | 0.36050 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -11.17 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
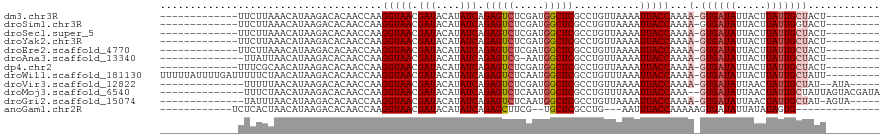
>dm3.chr3R 14541350 90 + 27905053 -------GAAAAGCAUUAUGAUUUGAAA---------GUUUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA -------.....((....((((.((...---------((.(((((....))))).)).((....)).......)).))))(((((.....)))))))......... ( -16.20, z-score = -1.12, R) >droSim1.chr3R 20572683 90 - 27517382 ------GAAAAGCAUUAUGAGUUGAAA----------GUUUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA ------.....((..((((.((((...----------((.(((((....))))).)).((....))..)))).))))...(((((.....)))))))......... ( -17.20, z-score = -1.17, R) >droSec1.super_5 507119 90 - 5866729 ------GAAAAGCAUUAUGAGUUGAAA----------GUUUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA ------.....((..((((.((((...----------((.(((((....))))).)).((....))..)))).))))...(((((.....)))))))......... ( -17.20, z-score = -1.17, R) >droYak2.chr3R 3198849 90 + 28832112 ------UGAAAACAUUAUGAGUUGAAC----------GUUUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA ------....((((.((((.((((...----------((.(((((....))))).)).((....))..)))).))))...(((((.....)))))...)))).... ( -16.80, z-score = -0.99, R) >droEre2.scaffold_4770 10654025 90 - 17746568 ------GAAAAACAUAAUGAUUUGAAA----------GUUUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA ------....(((..............----------((.(((((....))))).)).....((((...(((....))).(((((.....)))))))))))).... ( -16.00, z-score = -1.52, R) >droAna3.scaffold_13340 17099121 90 - 23697760 ----AAGAAAAACAUUUUCGGUUCAAA-----------AAUUAUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCG-AAUGGCUCGCCUGUUAAAA ----..((((.....))))((((....-----------..((((....)))).....)))).((((...(((....))).(((((.-...)))))))))....... ( -14.20, z-score = -1.33, R) >dp4.chr2 20192566 96 - 30794189 GAAGAGGGACGUGAAUUUGAGUUUAGUG---------UUUUCGC-AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA .((.(((..(((...((((.(((..(((---------((((...-.....))))))))))))))...)))..........(((((.....))))).))).)).... ( -22.60, z-score = -0.97, R) >droVir3.scaffold_12822 864886 90 + 4096053 -----ACAAUUAAAUUUUACGUU--AAU---------UGAUUUUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA -----.(((((((........))--)))---------))..(((((((....(.......).((((...(((....))).(((((.....)))))))))))))))) ( -16.40, z-score = -1.92, R) >droMoj3.scaffold_6540 33531134 100 + 34148556 ------GAAAACGAUUUUUGAUUUCAAGUAUUUACUGAUAUUUCUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUUAAA ------..(((((.(((.((.((..(((((((....)))))))..)))).))).).......((((...(((....))).(((((.....)))))))))))))... ( -15.30, z-score = -0.55, R) >droGri2.scaffold_15074 4815460 88 - 7742996 ---------GAAAAGUUUUCGUUGAAAU---------AUAUAUUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUAAAA ---------.....(((((.((((((..---------.....))))))..))))).......((((...(((....))).(((((.....)))))))))....... ( -16.10, z-score = -1.88, R) >anoGam1.chr2R 20738626 83 + 62725911 --------ACACGACUUC-AGUUCAGCA---------UUCUCACUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGCUUCGU--GCUCGCCUGAAU--- --------..........-.(((((((.---------..........................((((........)))).((((.....--))))).))))))--- ( -12.70, z-score = -0.51, R) >consensus ______GAAAAACAUUUUGAGUUGAAA__________GUUUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAA ..............................................................((((...(((....))).(((((.....)))))))))....... (-11.17 = -11.18 + 0.01)
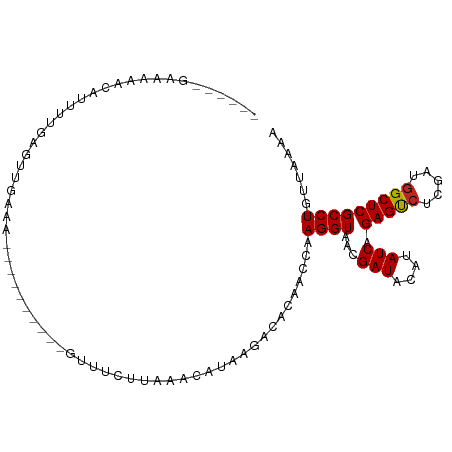
| Location | 14,541,373 – 14,541,470 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Shannon entropy | 0.25509 |
| G+C content | 0.34314 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.63 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
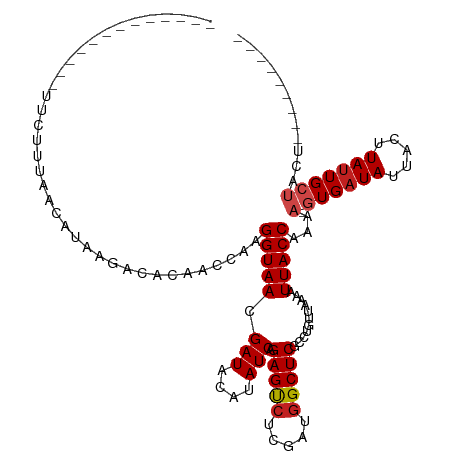
>dm3.chr3R 14541373 97 + 27905053 -------------UUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU--------- -------------.(((((....))))).........(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...--------- ( -18.50, z-score = -2.05, R) >droSim1.chr3R 20572706 97 - 27517382 -------------UUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGGUACU--------- -------------.(((((....)))))........((((...(((....))).(((((.....)))))))))........((((((((-(((....))))).))))))..--------- ( -21.10, z-score = -2.52, R) >droSec1.super_5 507142 97 - 5866729 -------------UUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU--------- -------------.(((((....))))).........(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...--------- ( -18.50, z-score = -2.05, R) >droYak2.chr3R 3198872 97 + 28832112 -------------UUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU--------- -------------.(((((....))))).........(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...--------- ( -18.50, z-score = -2.05, R) >droEre2.scaffold_4770 10654048 97 - 17746568 -------------UUCUUAAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU--------- -------------.(((((....))))).........(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...--------- ( -18.50, z-score = -2.05, R) >droAna3.scaffold_13340 17099146 95 - 23697760 --------------UUAUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCG-AAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU--------- --------------.......................(((((.(((....))).(((((.-...)))))...........)))))...(-(..(((.....)))..))...--------- ( -16.00, z-score = -1.37, R) >dp4.chr2 20192595 97 - 30794189 -------------UUUCGCAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU--------- -------------....(((..(((((.........((((...(((....))).(((((.....)))))))))..(((.(((((.....-))))).)))))))))))....--------- ( -18.80, z-score = -1.80, R) >droWil1.scaffold_181130 2570228 110 + 16660200 UUUUUAUUUUGAUUUUUCUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUUAAAUUACCAAAA-GUGAUAUUACUUAUUGCUAUU--------- .((((((.((((....)).)).)))))).........(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...--------- ( -17.70, z-score = -1.20, R) >droVir3.scaffold_12822 864910 98 + 4096053 --------------UUUUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUAACUAUUGCUAU--AUA----- --------------......................((((...(((....))).(((((.....)))))))))(((((.(((((.....-))))).))))).........--...----- ( -17.30, z-score = -1.56, R) >droMoj3.scaffold_6540 33531168 104 + 34148556 --------------UUUCUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUUAAAUUACCAAA--GUGAUAUUAACUAUUGCUAUUAGUACGAUA --------------...((((................(((((.(((....))).(((((.....)))))...........)))))..(--(..(((.....)))..)).))))....... ( -16.80, z-score = -0.68, R) >droGri2.scaffold_15074 4815482 99 - 7742996 --------------UAUUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUAACUAUUGCUAU-AGUA----- --------------.............((.......((((...(((....))).(((((.....)))))))))(((((.(((((.....-))))).))))).........-.)).----- ( -17.40, z-score = -1.45, R) >anoGam1.chr2R 20738646 89 + 62725911 ------------UCUCACUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGCUUCG--UGCUCGCCUG---AAUUUACCAAAAAGUGAUAUUAUAUAGUG-------------- ------------...(((((..((((..(((......(((((.(((....))).((((....--.)))).....---...))))).....)))...)))).)))))-------------- ( -16.60, z-score = -1.95, R) >consensus _____________UUCUUUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA_GUGAUAUUACUUAUUGCUACU_________ .....................................(((((.(((....))).(((((.....)))))...........))))).....((((((.....))))))............. (-13.38 = -13.63 + 0.26)
| Location | 14,541,379 – 14,541,470 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.72 |
| Shannon entropy | 0.26130 |
| G+C content | 0.35283 |
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.63 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14541379 91 + 27905053 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU---------------- ------------..................(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.90, z-score = -1.67, R) >droSim1.chr3R 20572712 91 - 27517382 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGGUACU---------------- ------------.................((((...(((....))).(((((.....)))))))))........((((((((-(((....))))).))))))..---------------- ( -19.50, z-score = -2.11, R) >droSec1.super_5 507148 91 - 5866729 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU---------------- ------------..................(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.90, z-score = -1.67, R) >droYak2.chr3R 3198878 91 + 28832112 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU---------------- ------------..................(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.90, z-score = -1.67, R) >droEre2.scaffold_4770 10654054 91 - 17746568 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU---------------- ------------..................(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.90, z-score = -1.67, R) >droAna3.scaffold_13340 17099151 90 - 23697760 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCG-AAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU---------------- ------------..................(((((.(((....))).(((((.-...)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.00, z-score = -1.57, R) >dp4.chr2 20192601 91 - 30794189 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUACUUAUUGCUACU---------------- ------------..................(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.90, z-score = -1.67, R) >droWil1.scaffold_181130 2570235 103 + 16660200 UUUGAUUUUUCUAACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUUAAAUUACCAAAA-GUGAUAUUACUUAUUGCUAUU---------------- ..............................(((((.(((....))).(((((.....)))))...........)))))...(-(..(((.....)))..))...---------------- ( -16.90, z-score = -1.09, R) >droVir3.scaffold_12822 864915 100 + 4096053 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUAACUAUUGCUAUA-UACAUGUAG------ ------------.((((............((((...(((....))).(((((.....)))))))))(((((.(((((.....-))))).)))))..........-...))))..------ ( -19.10, z-score = -1.66, R) >droMoj3.scaffold_6540 33531173 106 + 34148556 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUUAAAUUACCAAA--GUGAUAUUAACUAUUGCUAUUAGUACGAUAUAUGUAC ------------.(((((............(((((.(((....))).(((((.....)))))...........)))))..(--(..(((.....)))..))............))))).. ( -19.30, z-score = -1.03, R) >droGri2.scaffold_15074 4815487 101 - 7742996 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCAAUGGCUCGCCUGUUAAAAUUACCAAAA-GUGAUAUUAACUAUUGCUAUAGUAUACAUAA------ ------------........((.......((((...(((....))).(((((.....)))))))))(((((.(((((.....-))))).)))))..........))........------ ( -17.40, z-score = -1.43, R) >anoGam1.chr2R 20738653 82 + 62725911 ------------AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGCUUCG--UGCUCGCCUG---AAUUUACCAAAAAGUGAUAUUAUAUAGUG--------------------- ------------.........(((.....((((...(((....))).((((....--.)))))))).---...((((......)))).........)))--------------------- ( -13.40, z-score = -1.18, R) >consensus ____________AACAUAAGACACAACCAAGGUAACGAUACAUAUCAGAGUCUCGAUGGCUCGCCUGUUAAAAUUACCAAAA_GUGAUAUUACUUAUUGCUACU________________ ..............................(((((.(((....))).(((((.....)))))...........))))).....((((((.....)))))).................... (-13.38 = -13.63 + 0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:51 2011