
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,537,030 – 14,537,122 |
| Length | 92 |
| Max. P | 0.616980 |

| Location | 14,537,030 – 14,537,122 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.37361 |
| G+C content | 0.47639 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -13.55 |
| Energy contribution | -13.49 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14537030 92 - 27905053 UAUGGCAUACACCAUCCCCAUCCAUACAAAUUCAGAGCCG-CCCCUUAUGAGGCUUUGGUGUACGCGAGACGUAACAAAAACAUUUCGUUUUA ....((.(((((((....................(((((.-..........)))))))))))).))((((((..............)))))). ( -19.19, z-score = -1.38, R) >droSim1.chr3R 20568405 92 + 27517382 UGUGGCAUACACCAUCCCCAUCCAUCCAAAUUCAGAGCCG-CCCCUUAUGAGGCUUUGGUGUACGCGAGACGUAACAAAAACAUUUCGUUUUA ....((.(((((((....................(((((.-..........)))))))))))).))((((((..............)))))). ( -19.19, z-score = -0.92, R) >droSec1.super_5 502848 92 + 5866729 UGUGGCGUACACCAUCCCCAUCCAAACAAAUUCAGAGCCG-CCCCUUAUGAGGCUUUGGUGUACGCGAGACGUAACAAAAACAUUUCGUUUUA ....((((((((((....................(((((.-..........)))))))))))))))((((((..............)))))). ( -24.89, z-score = -2.50, R) >droYak2.chr3R 3194705 92 - 28832112 UAUGCCAUAAACCAUCCCCAUCCAUCCAAAUUCAGAGCCG-CCCCUUAUGAGGCUUUGGUGUGUACGAGACGUAGCAAAAACAUUUCGUUUUA ..(((.....................((...((((((((.-..........))))))))..))((((...)))))))(((((.....))))). ( -15.50, z-score = -0.52, R) >droEre2.scaffold_4770 10649802 80 + 17746568 ------------CAUCUCCAUCCAUCCAAAUGCAGAGCCG-CCCCUUAUGAGGCUUUGGUGUGUGCGAGACGUAACAAAAACAUUUCGUUUUA ------------..((((((..(((.......(((((((.-..........))))))))))..)).)))).......(((((.....))))). ( -17.41, z-score = -0.96, R) >droAna3.scaffold_13340 17095019 88 + 23697760 ----CCCCACCACAUCCCUUACCAUCUAAGUCCAGAACCU-CCCCUUAUGAGGCUUUGGCGUACACGAGACGUAACAAAAACAUUUCGUUUUA ----.........................(.(((((.(((-(.......)))).))))))....((((((.((.......)).)))))).... ( -14.90, z-score = -1.74, R) >dp4.chr2 20188601 81 + 30794189 ------------CCCCUGCCCCCUGCCCAGGGCAGAGCCGCCCCCUCAUCGGGCUUUGGUGUAUGCGAGACGUAACAAAAACAUUUCGUUUUA ------------...((((((........)))))).(((((((.......))))...)))....((((((.((.......)).)))))).... ( -23.80, z-score = -0.93, R) >consensus U_UGGCAUACACCAUCCCCAUCCAUCCAAAUUCAGAGCCG_CCCCUUAUGAGGCUUUGGUGUACGCGAGACGUAACAAAAACAUUUCGUUUUA ................................(((((((............)))))))......((((((.((.......)).)))))).... (-13.55 = -13.49 + -0.06)

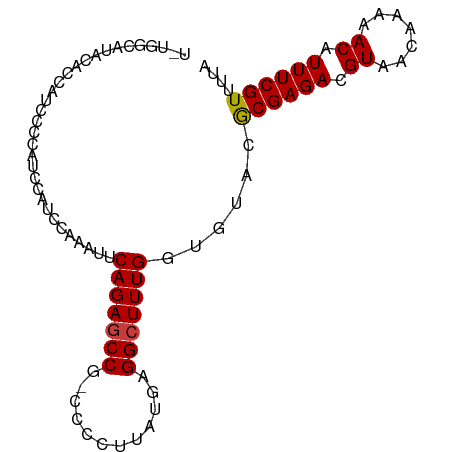
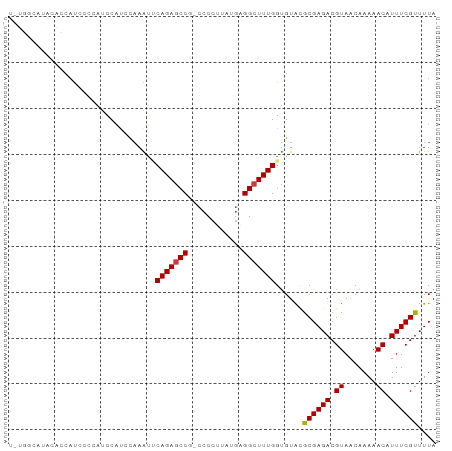
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:48 2011