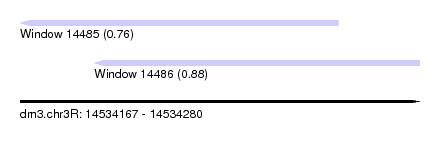
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,534,167 – 14,534,280 |
| Length | 113 |
| Max. P | 0.880742 |

| Location | 14,534,167 – 14,534,257 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.10 |
| Shannon entropy | 0.69685 |
| G+C content | 0.51317 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -9.02 |
| Energy contribution | -8.09 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
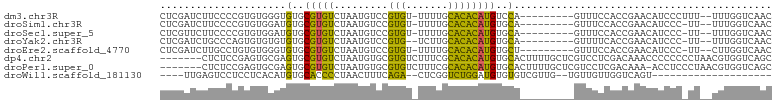
>dm3.chr3R 14534167 90 - 27905053 CUCGAUCUUCCCCGUGUGGGUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUCCA---------GUUUCCACCGAACAUCCCUUU--UUUGGUCAAC ...((((........(.((((((..(((.....((..(((((-........)))))..))---------.....)))...)))))))...--...))))... ( -19.84, z-score = -0.64, R) >droSim1.chr3R 20565636 89 + 27517382 CUCGAUCUUCCCCGUGUGGAUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUGCA---------GUUUCCACCGAACAUCCC-UU--UUUGGUCAAC ...((((........(.((((((((((.....))))..(((.-..((((((....)))))---------)....)))...)))))))-..--...))))... ( -21.12, z-score = -1.02, R) >droSec1.super_5 500132 89 + 5866729 CUCGUUCUUCCCCGUGUGGAUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUGCA---------GUUUCCACCGAACAUCCC-UU--UUUGGUCAAC ............((.(((((((..(((((.........((((-....)))))))))..))---------...)))))))......((-..--...))..... ( -20.50, z-score = -1.04, R) >droYak2.chr3R 3191829 88 - 28832112 CUCGAUCUGCCCAGUGUGUGUGUGCGUGUCUAAUGUCCGUG--UCUUGCACACAUGUGCA---------GUUUUCACCGAACAUCCC-UU--UUUGGUCAAC ...((.((((...(((((((((..((.(.(....).)))..--)...))))))))..)))---------)...))((((((......-..--)))))).... ( -24.60, z-score = -1.85, R) >droEre2.scaffold_4770 10647156 89 + 17746568 CUCGAUCUUGCCUGUGUGGGUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUGCU---------GUUUCCACCGAACAUCCC-UU--CUUGGUCAAC ...((((......(.(((((.(..(((((.........((((-....)))))))))..).---------...))))))(((......-))--)..))))... ( -19.90, z-score = -0.11, R) >dp4.chr2 20185753 95 + 30794189 -------CUCUCCGAGUGCGAGUGCGUGUCUAAUGUGCGUGUCUUUCGCACACAUGUGCACUUUUGCUCGUCCUCGACAAACCCCCCCCUAACGUGGUCAGC -------.....((((..((((((.((((...(((((.(((.....))).)))))..))))...))))))..))))..........(((....).))..... ( -29.30, z-score = -2.52, R) >droPer1.super_0 9871491 94 - 11822988 -------CUCUCCGAGUGCGAGUGCGUGUCUAAUGUGCGUGUCUUUCGCACACAUGUGCACUUUUGCUCGUCCUCGACAAA-ACCUCCCUAACGUGGUCAGC -------.....((((..((((((.((((...(((((.(((.....))).)))))..))))...))))))..)))).....-....(((....).))..... ( -29.10, z-score = -2.22, R) >droWil1.scaffold_181130 2561728 74 - 16660200 ----UUGAGUCCUCCUCACAUGUGCACCCCUAACUUUCAGA--CUCGGUCUGGAUGUGUGUCGUUG--UGUUGUUGGUCAGU-------------------- ----....(.((..(.((((...((((.....((.((((((--(...))))))).))))))...))--))..)..)).)...-------------------- ( -17.60, z-score = 0.03, R) >consensus CUCGAUCUUCCCCGUGUGGGUGUGCGUGUCUAAUGUCCGUGU_UUUUGCACACAUGUGCA_________GUUUCCACCGAACAUCCC_UU__UUUGGUCAAC .....................(..(((((.........((((.....)))))))))..)........................................... ( -9.02 = -8.09 + -0.94)

| Location | 14,534,188 – 14,534,280 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 60.39 |
| Shannon entropy | 0.75032 |
| G+C content | 0.50397 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -6.45 |
| Energy contribution | -6.42 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14534188 92 - 27905053 -------GAGCUCCAAAUGGUUUCCUCUUUCUCGAUCUUCCCCGUGUGGGUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUCCAGUUUCCACCG------ -------(((..((....))....)))...............((.(((((((..(((((.........((((-....)))))))))..))...)))))))------ ( -17.80, z-score = -0.39, R) >droSim1.chr3R 20565656 92 + 27517382 -------GAGCUCCAAAUGGUUUCCUCUUUCUCGAUCUUCCCCGUGUGGAUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUGCAGUUUCCACCG------ -------(((..((....))....)))...............((.(((((((..(((((.........((((-....)))))))))..))...)))))))------ ( -21.70, z-score = -1.71, R) >droSec1.super_5 500152 92 + 5866729 -------GAGCCCCAAAUGGUUUCCUCUUUCUCGUUCUUCCCCGUGUGGAUGUGCGUGUCUAAUGUCCGUGU-UUUUGCACACAUGUGCAGUUUCCACCG------ -------(((((......)))))...................((.(((((((..(((((.........((((-....)))))))))..))...)))))))------ ( -21.50, z-score = -2.08, R) >droYak2.chr3R 3191849 91 - 28832112 -------GAGCCCCAAAUGGUUUCCUCUUUCUCGAUCUGCCCAGUGUGUGUGUGCGUGUCUAAUGUCCGUG--UCUUGCACACAUGUGCAGUUUUCACCG------ -------(((((......)))))..........((.((((...(((((((((..((.(.(....).)))..--)...))))))))..))))...))....------ ( -23.70, z-score = -2.23, R) >droAna3.scaffold_13340 17091967 82 + 23697760 -------GAGUGCCAAAUGGCUUCCCCCACGC---------UCUAAUAUCC-UCCGUGUCUAAUAUCCCUGU-AUUCGCACACAUGGAGACUUUCCAUCG------ -------(((((.....(((......))))))---------)).......(-((((((((.(((((....))-))).)...))))))))...........------ ( -16.40, z-score = -2.04, R) >dp4.chr2 20185779 90 + 30794189 -------GACUCUCAAAAUGUUCUUUUUCUCU---------CCGAGUGCGAGUGCGUGUCUAAUGUGCGUGUCUUUCGCACACAUGUGCACUUUUGCUCGUCCUCG -------.........................---------.((((..((((((.((((...(((((.(((.....))).)))))..))))...))))))..)))) ( -26.70, z-score = -3.40, R) >droPer1.super_0 9871516 90 - 11822988 -------GACUCUCAAAAUGUUCUUUUUCUCU---------CCGAGUGCGAGUGCGUGUCUAAUGUGCGUGUCUUUCGCACACAUGUGCACUUUUGCUCGUCCUCG -------.........................---------.((((..((((((.((((...(((((.(((.....))).)))))..))))...))))))..)))) ( -26.70, z-score = -3.40, R) >droWil1.scaffold_181130 2561741 95 - 16660200 GGAAGUGCGGUGCGGAAUGGUGGAAGUGUUUCUUUUGAGUCCUCCUCACAUGUGCACCCCUAACUUUCAGACUCGGUCUGGAUGUGUGUCGUUGU----------- .((.(..(((((((..((((.(((.(...(((....))).).))).).))).)))))).......(((((((...))))))).)..).)).....----------- ( -25.40, z-score = 0.39, R) >consensus _______GAGCCCCAAAUGGUUUCCUCUUUCUCG_UCU_CCCCGUGUGCGUGUGCGUGUCUAAUGUCCGUGU_UUUUGCACACAUGUGCAGUUUCCACCG______ ..............................................((((((((.((((..................)))).))))))))................ ( -6.45 = -6.42 + -0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:46 2011