
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,510,509 – 14,510,611 |
| Length | 102 |
| Max. P | 0.727094 |

| Location | 14,510,509 – 14,510,611 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.31 |
| Shannon entropy | 0.71906 |
| G+C content | 0.39878 |
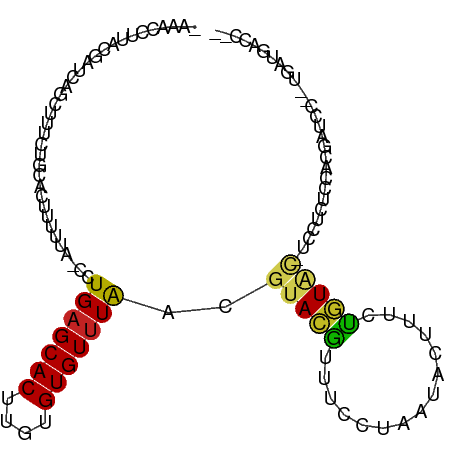
| Mean single sequence MFE | -14.47 |
| Consensus MFE | -6.20 |
| Energy contribution | -6.21 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
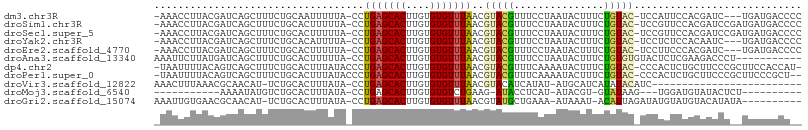
>dm3.chr3R 14510509 102 - 27905053 -AAACCUUACGAUCAGCUUUCUGCAAUUUUUA-CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUAC-UCCAUUCCACGAUC---UGAUGACCCC -.........((((.((.....))........-..(((((((....)))))))..(((((...............)))))-..........))))---.......... ( -14.56, z-score = -1.25, R) >droSim1.chr3R 20540913 105 + 27517382 -AAACCUUACGAUCAGCUUUCUGCACUUUUUA-CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUAC-UCCGUUCCACGAUCCGAUGAUGACCCC -.........((((.((.....))........-..(((((((....)))))))..(((((...............)))))-..........))))............. ( -14.16, z-score = -0.36, R) >droSec1.super_5 476393 105 + 5866729 -AAACCUUACGAUCAGCUUUCUGCACUUUUUA-CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUAC-UCCGUUCCACGAUCCGAUGAUGACCCC -.........((((.((.....))........-..(((((((....)))))))..(((((...............)))))-..........))))............. ( -14.16, z-score = -0.36, R) >droYak2.chr3R 3162896 102 - 28832112 -AAACCUUACGAUCAGCUUUCUGCACAUUUUA-CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUAC-UCCUCUCCACAAUC---UGAUGACCCC -..........(((((................-..(((((((....)))))))..(((((...............)))))-.............)---))))...... ( -12.66, z-score = -0.76, R) >droEre2.scaffold_4770 10623402 102 + 17746568 -AAACCUUACGAUCAGCUUUCUGCACUUUUUA-CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUAC-UCCUUCCCACGAUC---UGAUGACCCC -.........((((.((.....))........-..(((((((....)))))))..(((((...............)))))-..........))))---.......... ( -14.56, z-score = -1.57, R) >droAna3.scaffold_13340 17068640 96 + 23697760 AAAUUCUUAUGAUCAGCUUUCUGCACUUUUUA-CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUGUGUACUCUCGAAGACCCU----------- ....((((.(((...((.....))........-..(((((((....)))))))..((((........((((.....))))))))..)))))))....----------- ( -13.30, z-score = -0.12, R) >dp4.chr2 20162317 105 + 30794189 -UAAUUUUACAGUCAGCUUUCUGCACUUUAUACCCUGAGCACUUGUGUGUUUAACGUACGUUUCAAAAUACUUUCUGUAC-CCCACUCUGCUUCCCGCUUCCACCAU- -......(((((...((.....))......(((..(((((((....)))))))..)))................))))).-........((.....)).........- ( -13.90, z-score = -1.23, R) >droPer1.super_0 9848445 104 - 11822988 -UAAUUUUACAGUCAGCUUUCUGCACUUUAUACCCUGAGCACUUGUGUGUUUAACGUACGUUUCAAAAUACUUUCUGUAC-CCCACUCUGCUUCCCGCUUCCCGCU-- -......(((((...((.....))......(((..(((((((....)))))))..)))................))))).-...............((.....)).-- ( -14.20, z-score = -1.09, R) >droVir3.scaffold_12822 830731 80 - 4096053 AAACUUUAAACGCAACAU-UCUGCACUUUAUA-CCUGAGCACUUGUGUGUUUAACGUACAUCAUAU-AUGCAUCAUAUACAUC------------------------- ..((.(((((((((.((.-..(((.(......-...).)))..))))))))))).))......(((-(((...))))))....------------------------- ( -13.50, z-score = -1.60, R) >droMoj3.scaffold_6540 33492037 80 - 34148556 -----------AAAAUAUGUCUGCACUUUAUA-CCUGAGCACUUGUGUGUCUGAAG-AUACCUCAU-AUACGU-GUAUAAG---UGGAUGUAUACUCU---------- -----------...(((((((..(...(((((-(.((......((.(((((....)-))))..)).-...)).-)))))))---..))))))).....---------- ( -16.10, z-score = -0.70, R) >droGri2.scaffold_15074 4781722 94 + 7742996 AAAUUGUGAACGCAACAU-UCUGCACUUUAUA-CCUGAGCACUUGUGUGUUUAACGUAUGCUGAAA-AUAAAU-ACAUUAGAUAUGUAUGUACAUAUA---------- ....((((.(((((....-..))).(..((((-(.(((((((....)))))))..)))))..)...-....((-((((.....)))))))).))))..---------- ( -18.10, z-score = -0.62, R) >consensus _AAACCUUACGAUCAGCUUUCUGCACUUUUUA_CCUGAGCACUUGUGUGUUUAACGUACGUUUCCUAAUACUUUCUGUAC_UCCUCUCCACGAUCC__UGAUGACC__ ...................................(((((((....)))))))....................................................... ( -6.20 = -6.21 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:44 2011