
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,505,857 – 14,505,962 |
| Length | 105 |
| Max. P | 0.743797 |

| Location | 14,505,857 – 14,505,962 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.87 |
| Shannon entropy | 0.78593 |
| G+C content | 0.44950 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -8.43 |
| Energy contribution | -7.91 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
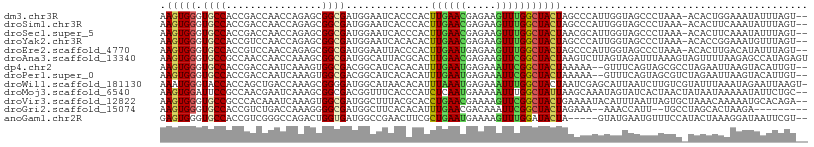
>dm3.chr3R 14505857 105 + 27905053 AAGUGGGUGCCACCGACCAACCAGAGCGGCGAUGGAAUCACCCACUUGAACGAGAAGUUUGGCUACUAGCCCAUUGGUAGCCCUAAA-ACACUGGAAAUAUUUAGU-- ((((((((((((.((.((.........)))).)))...)))))))))((((.....))))(((((((((....))))))))).....-..((((((....))))))-- ( -34.20, z-score = -2.79, R) >droSim1.chr3R 20536216 105 - 27517382 AAGUGGGUGCCACCGACCAACCAGAGCGGCGAUGGAAUCACCCACUUGAACGAGAAGUUUGGCUACUAGCCCAUUGGUAGCCCUAAA-ACACUUCAAAUAUUUAGU-- ((((((((((((.((.((.........)))).)))...)))))))))......(((((..(((((((((....))))))))).....-..)))))...........-- ( -34.30, z-score = -3.23, R) >droSec1.super_5 471669 105 - 5866729 AAGUGGGUGCCACCGACCAACCAGAGCGGCGAUGGAAUCACCCACUUGAACGAGAAGUUUGGCUACUAACGCAUUGGUAGCCCUAAA-ACACUUCAAAUAUUUAGU-- ((((((((((((.((.((.........)))).)))...)))))))))......(((((..(((((((((....))))))))).....-..)))))...........-- ( -34.60, z-score = -3.49, R) >droYak2.chr3R 3158306 105 + 28832112 AAGUGGGUGCCACCGUCCAACCAGAGCGGCGAUGGAAUCACACACUUGAACGAGAAGUUUGGCUACUAGCCCAUUGGUAGCCCUAAA-ACACCGGAAAUGUUUAGU-- (((((.(((...(((((...((.....)).)))))...))).)))))((((.....))))(((((((((....)))))))))...((-(((.......)))))...-- ( -27.90, z-score = -0.24, R) >droEre2.scaffold_4770 10618934 105 - 17746568 AAGUGGGUGCCACCGUCCAACCAGAGCGGCGAUGGAAUUACCCACUUGAAUGAGAAGUUUGGCUACUAGCCCAUUGGUAGCCCUAAA-ACACUUGACAUAUUUAGU-- (((((((((...(((((...((.....)).)))))...))))))))).........(((((((((((((....)))))))))...))-))................-- ( -32.30, z-score = -2.28, R) >droAna3.scaffold_13340 17064137 108 - 23697760 AAGUGGGUGCCGCCAACCAACCAAAGCGGCGAUGGCAUUACGCACUUGAACGAGAAGUUCGGCUACUAAGUCUUAGUAGAUUUAAAGUAGUUUUAAGAGCCAUAGAGU .......((((((............))))))(((((...........((((.....))))(((((((((((((....))))))..)))))))......)))))..... ( -29.70, z-score = -1.43, R) >dp4.chr2 20157810 104 - 30794189 AAGUGGGUGCCACCGACCAAUCAAAGUGGCGACGGCAUCACACAUUUGAAUGAGAAAUUCGGCUACUAAAAA--GUUUCAGUAGCGCCUAGAAUUAAGUACAUUGU-- ..(((((((((..((.(((.......)))))..)))))).)))...(((((.....)))))((((((.((..--..)).)))))).....................-- ( -26.90, z-score = -1.54, R) >droPer1.super_0 9843974 104 + 11822988 AAGUGGGUGCCACCGACCAAUCAAAGUGGCGACGGCAUCACACAUUUGAAUGAGAAAUUCGGCUACUAAAAA--GUUUCAGUAGCGUCUAGAAUUAAGUACAUUGU-- ..(((((((((..((.(((.......)))))..)))))).)))...(((((.....)))))((((((.((..--..)).)))))).....................-- ( -26.90, z-score = -1.65, R) >droWil1.scaffold_181130 2471576 107 + 16660200 AAAUGGGUACCACCAGCUGACCAAAGCGGGGAUGGCAUAACACAUUUAAAUGAGAAAUUUGGCUACUAAUCGAGCAUUAAUCUUGUCGUAUUUAAAUAGAAUUAAGU- (((((.((.(((((.(((......)))..)).))).))....)))))..((((.(((((..(((........)))...))).)).))))..................- ( -17.30, z-score = 0.66, R) >droMoj3.scaffold_6540 33486946 106 + 34148556 AAGUGGAUUCCGCCAACGAAUCAAAGCGGCGACGGUUUCACCCAUCUCAAUGAAAAAUUUGGCUAUUAAGCAAAUAGUAUCACUAACUAUAAUAAAAAUAUUCUGC-- .((((((..(((.(..((........))..).)))..))......................((((((.....))))))..))))......................-- ( -14.50, z-score = 0.57, R) >droVir3.scaffold_12822 825974 106 + 4096053 AAGUGGGUGCCGCCCACAAAUCAAAGUGGCGAUGGCUUUACGCACCUGAACGAAAAGUUCGGCUACUGAAAAUACAUUUAAUUAGUGCUAAACAAAAAUGCACAGA-- ..(((((.....))))).......((((((....((.....))...(((((.....))))))))))).................((((...........))))...-- ( -27.60, z-score = -1.54, R) >droGri2.scaffold_15074 4775813 95 - 7742996 AAGUGGGUGCCACCGUCUGACCAAAGGGGCGAUGGCUUCACACAUUUGAACGACAAAUUCGGCUACUAGAAA--AAACCAUU--UGCCUAGCACUAAGA--------- ..(((((.((((.(((((........))))).))))))))).........(((.....)))((((...(((.--......))--)...)))).......--------- ( -21.40, z-score = 0.06, R) >anoGam1.chr2R 59920955 101 + 62725911 GAGUGGGUGCCACCGUCGGGCCAGACUGGUGAUGGCCGAACUUCGCUGAAUGAAAAGUUUGGAUACUA-----GUAUGAAUGUUUCCAUACUAAAGGAUAAUUCGU-- ((((.....((.(((((..(((.....))))))))((((((((..(.....)..))))))))....((-----(((((........)))))))..))...))))..-- ( -28.50, z-score = -1.16, R) >consensus AAGUGGGUGCCACCGACCAACCAAAGCGGCGAUGGCAUCACACACUUGAACGAGAAGUUUGGCUACUAACCCAUUGGUAACCCUAAA_AAACUUAAAAUAUUUAGU__ .(((((.((((................))))..............((((((.....)))))))))))......................................... ( -8.43 = -7.91 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:42 2011