| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,913,491 – 7,913,613 |
| Length | 122 |
| Max. P | 0.830873 |

| Location | 7,913,491 – 7,913,583 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 91.29 |
| Shannon entropy | 0.16308 |
| G+C content | 0.46270 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -19.84 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
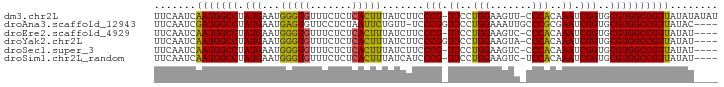
>dm3.chr2L 7913491 92 + 23011544 UUCAAUCAAUGGCCUAUGAAUGGGUGUUUCUCUCACUUUAUCUUCCCG-UUCCUGGAAGUU-CCCACAAAUCGGUGCGUGGCCGUUAUAUAUAU .......(((((((...(((((((....................))))-)))......((.-((........)).))..)))))))........ ( -20.45, z-score = -0.84, R) >droAna3.scaffold_12943 3646246 89 - 5039921 UUCAAUCGAUGGCCUAUGAAUGAGUGUUCCUCUAAUUCUGUU-UCCCGGUUCCUGGAAAUUGCCCGCGAAUCGGUGCGUGGCCGUUAUAC---- .......(((((((...(((((((.....)))..)))).((.-..(((((((.(((.......))).))))))).))..)))))))....---- ( -26.00, z-score = -1.86, R) >droEre2.scaffold_4929 16835591 88 + 26641161 UUCAAUCAAUGGCCUAUGAAUGGGUGUUUCUCUCACUUUAUCUUCCCG-UUCCUGGAAGUC-CCCACAAAUCGGUGCGUGGCCGUUAUAU---- .......(((((((...(((((((....................))))-)))......(..-((........))..)..)))))))....---- ( -20.95, z-score = -1.04, R) >droYak2.chr2L 17343214 89 - 22324452 UUCAAUCAAUGGCCUAUGAAUGGGUGUUUCUCUCACUUUAUCUUCCCGGUUCCUGGAAGUA-CCCACAAAUCGGUGCGUGGCCGUUAUAU---- .......(((((((.(((...(((((.......))))).......((((((..(((.....-.)))..))))))..))))))))))....---- ( -24.20, z-score = -1.52, R) >droSec1.super_3 3427877 88 + 7220098 UUCAAUCAAUGGCCUAUGAAUGGGUGUUUCUCUCACUUUAUCUUCCCG-UUCCUGGAAGUC-CCCACAAAUCGGUGCGUGGCCGUUAUAU---- .......(((((((...(((((((....................))))-)))......(..-((........))..)..)))))))....---- ( -20.95, z-score = -1.04, R) >droSim1.chr2L_random 411951 88 + 909653 UUCAAUCAAUGGCCUAUGAAUGGGUGUUUCUCUCACUUUAUCAUCCCG-UUCCUGGAAGUC-UCCACAAAUCGGUGCGUGGCCGUUAUAU---- .......(((((((.((((..(((((.......)))))..)))).(((-((..((((....-))))..)).))).....)))))))....---- ( -22.40, z-score = -1.65, R) >consensus UUCAAUCAAUGGCCUAUGAAUGGGUGUUUCUCUCACUUUAUCUUCCCG_UUCCUGGAAGUC_CCCACAAAUCGGUGCGUGGCCGUUAUAU____ .......(((((((.(((...(((((.......))))).......(((.((..(((.......)))..)).)))..))))))))))........ (-19.84 = -19.32 + -0.53)


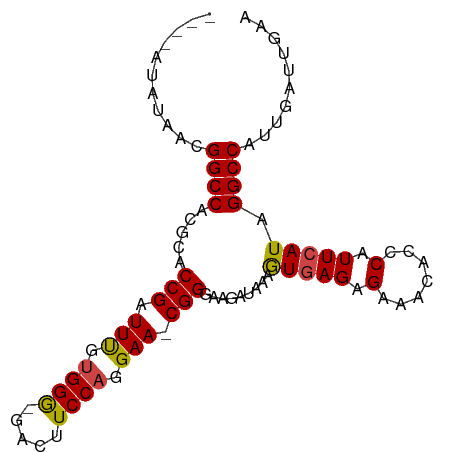
| Location | 7,913,491 – 7,913,583 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 91.29 |
| Shannon entropy | 0.16308 |
| G+C content | 0.46270 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7913491 92 - 23011544 AUAUAUAUAACGGCCACGCACCGAUUUGUGGG-AACUUCCAGGAA-CGGGAAGAUAAAGUGAGAGAAACACCCAUUCAUAGGCCAUUGAUUGAA ...........((((.....(((.(((.((((-....)))).)))-)))...((....(((.......)))....))...)))).......... ( -20.90, z-score = -1.20, R) >droAna3.scaffold_12943 3646246 89 + 5039921 ----GUAUAACGGCCACGCACCGAUUCGCGGGCAAUUUCCAGGAACCGGGA-AACAGAAUUAGAGGAACACUCAUUCAUAGGCCAUCGAUUGAA ----.......((((..((.(((.....)))))..(((((.(....).)))-))..((((..(((.....)))))))...)))).......... ( -24.20, z-score = -2.02, R) >droEre2.scaffold_4929 16835591 88 - 26641161 ----AUAUAACGGCCACGCACCGAUUUGUGGG-GACUUCCAGGAA-CGGGAAGAUAAAGUGAGAGAAACACCCAUUCAUAGGCCAUUGAUUGAA ----.......((((.((((......))))((-(.(((((.....-..))))).....((.......)).))).......)))).......... ( -22.60, z-score = -1.40, R) >droYak2.chr2L 17343214 89 + 22324452 ----AUAUAACGGCCACGCACCGAUUUGUGGG-UACUUCCAGGAACCGGGAAGAUAAAGUGAGAGAAACACCCAUUCAUAGGCCAUUGAUUGAA ----.......((((.((...))....(((((-(.(((((.(....).))))).....((.......)))))))).....)))).......... ( -22.40, z-score = -1.20, R) >droSec1.super_3 3427877 88 - 7220098 ----AUAUAACGGCCACGCACCGAUUUGUGGG-GACUUCCAGGAA-CGGGAAGAUAAAGUGAGAGAAACACCCAUUCAUAGGCCAUUGAUUGAA ----.......((((.((((......))))((-(.(((((.....-..))))).....((.......)).))).......)))).......... ( -22.60, z-score = -1.40, R) >droSim1.chr2L_random 411951 88 - 909653 ----AUAUAACGGCCACGCACCGAUUUGUGGA-GACUUCCAGGAA-CGGGAUGAUAAAGUGAGAGAAACACCCAUUCAUAGGCCAUUGAUUGAA ----.......((((.....(((.(((.(((.-.....))).)))-))).((((....(((.......)))....)))).)))).......... ( -23.30, z-score = -2.18, R) >consensus ____AUAUAACGGCCACGCACCGAUUUGUGGG_GACUUCCAGGAA_CGGGAAGAUAAAGUGAGAGAAACACCCAUUCAUAGGCCAUUGAUUGAA ...........((((.....(((.(((.((((.....)))).))).))).........(((((.(.......).))))).)))).......... (-20.10 = -20.02 + -0.08)


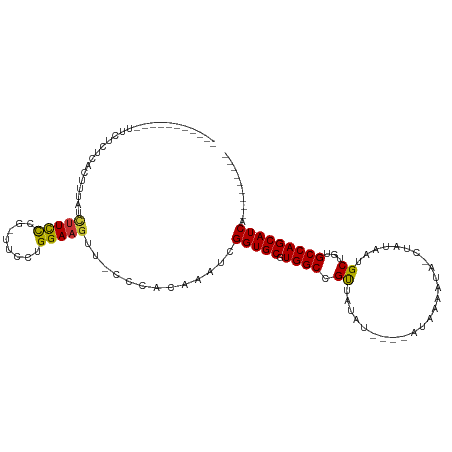
| Location | 7,913,518 – 7,913,613 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.00 |
| Shannon entropy | 0.49259 |
| G+C content | 0.45915 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -13.44 |
| Energy contribution | -12.97 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7913518 95 + 23011544 -----------UUCUCUCACUUUAUCUUCCCG-UUCCUGGAAGUU-CCCACAAAUCGGUGCGUGGCCGUUAUAUAUAUAUAAAAUA-CUAUAAUGCUGUGCCAGCAUCA--------- -----------..............(((((..-.....)))))..-..........(((((.((((((((((((((.......)))-.)))))))....))))))))).--------- ( -19.90, z-score = -2.05, R) >droPer1.super_1 3415301 110 - 10282868 ACAGACUCCAGACUCCACUCUCCGUUUUUUCUGUUCCUGGAAGCU----GGACAACGGUGCGUGGCGGCCUGUUG---UUGUCGUCGUUAU-AUGCUGUGCCAGCAUCAUCAUCAUCA .(((....((((....((.....))....))))...)))((.(((----((...(((((..((((((((......---.....))))))))-..))))).))))).)).......... ( -30.00, z-score = -0.09, R) >dp4.chr4_group3 6316788 110 - 11692001 ACAGACUCCAGACUCCACUCUCCGUUUUUUCUGUUCCUGGAAGCU----GGACAACGGUGCGUGGCGGCCUGUUG---UUGUCGUCGUUAU-AUGCUGUGCCAGCAUCAUCAUCAUCA .(((....((((....((.....))....))))...)))((.(((----((...(((((..((((((((......---.....))))))))-..))))).))))).)).......... ( -30.00, z-score = -0.09, R) >droAna3.scaffold_12943 3646273 93 - 5039921 -----------UCCUCUAAUUCUGU-UUCCCGGUUCCUGGAAAUUGCCCGCGAAUCGGUGCGUGGCCGUUAUAC----AUAAGAGAACUAUUAUGCUGUGCCAGCAUCA--------- -----------..((((......((-((((.(....).)))))).(((((((......)))).)))........----...)))).......((((((...))))))..--------- ( -23.50, z-score = -0.82, R) >droEre2.scaffold_4929 16835618 91 + 26641161 -----------UUCUCUCACUUUAUCUUCCCG-UUCCUGGAAGUC-CCCACAAAUCGGUGCGUGGCCGUUAUAU----AUAAAAUA-CUAUAAUGCUGUGCCAGCAUCA--------- -----------..............(((((..-.....)))))..-..........(((((.(((((((((((.----........-.)))))))....))))))))).--------- ( -19.40, z-score = -2.13, R) >droYak2.chr2L 17343241 92 - 22324452 -----------UUCUCUCACUUUAUCUUCCCGGUUCCUGGAAGUA-CCCACAAAUCGGUGCGUGGCCGUUAUAU----AUAAAAUA-CUAUAAUGCUGUGCCAGCAUCA--------- -----------..............(((((.(....).)))))..-..........(((((.(((((((((((.----........-.)))))))....))))))))).--------- ( -21.40, z-score = -1.93, R) >droSec1.super_3 3427904 91 + 7220098 -----------UUCUCUCACUUUAUCUUCCCG-UUCCUGGAAGUC-CCCACAAAUCGGUGCGUGGCCGUUAUAU----AUAAAAUA-CUAUAAUGCUGUGCCAGCAUCA--------- -----------..............(((((..-.....)))))..-..........(((((.(((((((((((.----........-.)))))))....))))))))).--------- ( -19.40, z-score = -2.13, R) >droSim1.chr2L_random 411978 91 + 909653 -----------UUCUCUCACUUUAUCAUCCCG-UUCCUGGAAGUC-UCCACAAAUCGGUGCGUGGCCGUUAUAU----AUAAAAUA-CUAUAAUGCUGUGCCAGCAUCA--------- -----------....................(-((..((((....-))))..))).(((((.(((((((((((.----........-.)))))))....))))))))).--------- ( -18.00, z-score = -1.55, R) >consensus ___________UUCUCUCACUUUAUCUUCCCG_UUCCUGGAAGUU_CCCACAAAUCGGUGCGUGGCCGUUAUAU____AUAAAAUA_CUAUAAUGCUGUGCCAGCAUCA_________ .........................(((((........))))).............(((((.((((.((.........................))...))))))))).......... (-13.44 = -12.97 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:42 2011