| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,454,186 – 14,454,276 |
| Length | 90 |
| Max. P | 0.998456 |

| Location | 14,454,186 – 14,454,276 |
|---|---|
| Length | 90 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.37947 |
| G+C content | 0.59069 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -28.11 |
| Energy contribution | -27.75 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.998456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
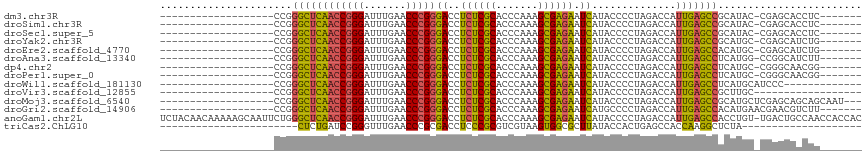
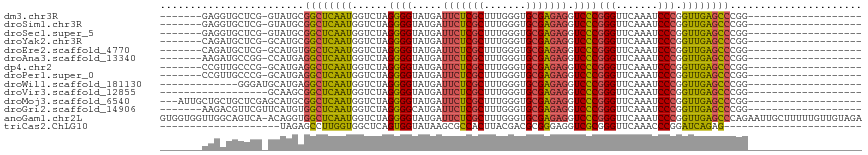
>dm3.chr3R 14454186 90 + 27905053 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCAUAC-CGAGCACCUC------- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))((....-...)).....------- ( -31.10, z-score = -3.62, R) >droSim1.chr3R 20484611 90 - 27517382 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCAUAC-CGAGCACCUC------- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))((....-...)).....------- ( -31.10, z-score = -3.62, R) >droSec1.super_5 420529 90 - 5866729 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCAUAC-CGAGCACCUC------- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))((....-...)).....------- ( -31.10, z-score = -3.62, R) >droYak2.chr3R 3097740 90 + 28832112 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCAUGC-CGAGCAUCUG------- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))((....-...)).....------- ( -31.10, z-score = -2.87, R) >droEre2.scaffold_4770 10566468 90 - 17746568 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCACAUGC-CGAGCAUCUG------- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))..((((-...))))...------- ( -30.90, z-score = -3.35, R) >droAna3.scaffold_13340 14034146 90 - 23697760 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCUCAUGG-CCGGCAUCUU------- -------------------(((((((((((((((.......)))))((..((((((.......)))))).))..............))))))))...))-..........------- ( -30.50, z-score = -1.80, R) >dp4.chr2 19076765 90 - 30794189 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCUCAUGC-CGGGCAACGG------- -------------------..(((((((((((((.......)))))((..((((((.......)))))).))..............))))))))....(-((.....)))------- ( -33.10, z-score = -2.48, R) >droPer1.super_0 8798177 90 + 11822988 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCUCAUGC-CGGGCAACGG------- -------------------..(((((((((((((.......)))))((..((((((.......)))))).))..............))))))))....(-((.....)))------- ( -33.10, z-score = -2.48, R) >droWil1.scaffold_181130 6117154 85 - 16660200 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCUCAUGCAUCCC------------- -------------------..(((((((((((((.......)))))((..((((((.......)))))).))..............))))))))..........------------- ( -30.40, z-score = -3.64, R) >droVir3.scaffold_12855 5775074 80 - 10161210 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCUUGC------------------ -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))......------------------ ( -30.10, z-score = -3.50, R) >droMoj3.scaffold_6540 25405382 95 - 34148556 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCAUGCUCGAGCAGCAGCAAU--- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............)))))))((.((((.....))))))...--- ( -35.90, z-score = -3.61, R) >droGri2.scaffold_14906 7216933 91 - 14172833 -------------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUGCCCCUAGACCAUUGAGCCACAUGAACGAACGUCUU------- -------------------...((((((((((((.......)))))((..((((((.......)))))).))..............))))))).................------- ( -30.10, z-score = -2.78, R) >anoGam1.chr2L 30714357 116 - 48795086 UCUACAACAAAAAGCAAUUCUGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCACCUGU-UGACUGCCAACCACCAC ............((((((....((((((((((((.......)))))((..((((((.......)))))).))..............)))))))....))-)).))............ ( -33.80, z-score = -3.85, R) >triCas2.ChLG10 77814 74 - 8806720 -----------------------CUCUGAUCCGGGUUUGAACCCGCGACCUCCCGCGUCGUAAGUGGCGCUUAUACCACUGAGCCACCAAGGCUCUA-------------------- -----------------------.........((((....))))((((((....).))))).(((((........)))))(((((.....)))))..-------------------- ( -24.80, z-score = -1.91, R) >consensus ___________________CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCAUGC_CGAGCACCUG_______ ......................((((((((((((.......)))))((..((((((.......)))))).))..............)))))))........................ (-28.11 = -27.75 + -0.36)
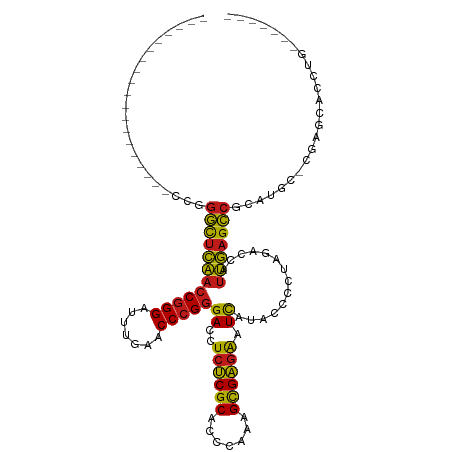
| Location | 14,454,186 – 14,454,276 |
|---|---|
| Length | 90 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.37947 |
| G+C content | 0.59069 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -30.02 |
| Energy contribution | -29.66 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
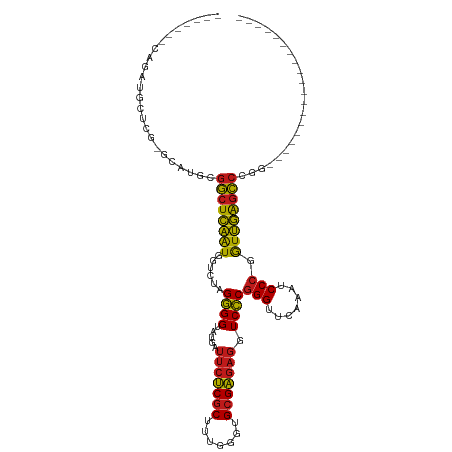
>dm3.chr3R 14454186 90 - 27905053 -------GAGGUGCUCG-GUAUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------(((.(((...-....))).)))..........(((.....(((((((.......)))))))..)))((((((((.......))))))))..------------------- ( -34.70, z-score = -0.94, R) >droSim1.chr3R 20484611 90 + 27517382 -------GAGGUGCUCG-GUAUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------(((.(((...-....))).)))..........(((.....(((((((.......)))))))..)))((((((((.......))))))))..------------------- ( -34.70, z-score = -0.94, R) >droSec1.super_5 420529 90 + 5866729 -------GAGGUGCUCG-GUAUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------(((.(((...-....))).)))..........(((.....(((((((.......)))))))..)))((((((((.......))))))))..------------------- ( -34.70, z-score = -0.94, R) >droYak2.chr3R 3097740 90 - 28832112 -------CAGAUGCUCG-GCAUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------...((((...-))))(.((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))))..------------------- ( -33.60, z-score = -0.48, R) >droEre2.scaffold_4770 10566468 90 + 17746568 -------CAGAUGCUCG-GCAUGUGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------...((((...-))))(.((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))))..------------------- ( -33.60, z-score = -0.78, R) >droAna3.scaffold_13340 14034146 90 + 23697760 -------AAGAUGCCGG-CCAUGAGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------...(((((((-((((........))))))....)))))(((((((((.......)))))).)))((((((((((.......))))))))))------------------- ( -36.90, z-score = -1.40, R) >dp4.chr2 19076765 90 + 30794189 -------CCGUUGCCCG-GCAUGAGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------((..(((((.-.....(((((....)))))..)))))....((((((.......))))))))..((((((((((.......))))))))))------------------- ( -35.10, z-score = -0.61, R) >droPer1.super_0 8798177 90 - 11822988 -------CCGUUGCCCG-GCAUGAGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------((..(((((.-.....(((((....)))))..)))))....((((((.......))))))))..((((((((((.......))))))))))------------------- ( -35.10, z-score = -0.61, R) >droWil1.scaffold_181130 6117154 85 + 16660200 -------------GGGAUGCAUGAGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------------(((((((...(((((....)))))....))....(((((((.......))))))))))))((((((((.......))))))))..------------------- ( -33.90, z-score = -1.33, R) >droVir3.scaffold_12855 5775074 80 + 10161210 ------------------GCAAGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- ------------------....(.((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))))..------------------- ( -32.30, z-score = -1.43, R) >droMoj3.scaffold_6540 25405382 95 + 34148556 ---AUUGCUGCUGCUCGAGCAUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- ---...((((((.....)))).))((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...------------------- ( -38.10, z-score = -1.20, R) >droGri2.scaffold_14906 7216933 91 + 14172833 -------AAGACGUUCGUUCAUGUGGCUCAAUGGUCUAGGGGCAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------------- -------..............((.((((((((......(((((....(((((((.......))))))))))))(((.......))).)))))))))).------------------- ( -35.10, z-score = -1.54, R) >anoGam1.chr2L 30714357 116 + 48795086 GUGGUGGUUGGCAGUCA-ACAGGUGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCAGAAUUGCUUUUUGUUGUAGA (..(..(..((((((..-....(.((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))))...))))))..)..)..)... ( -40.90, z-score = -1.42, R) >triCas2.ChLG10 77814 74 + 8806720 --------------------UAGAGCCUUGGUGGCUCAGUGGUAUAAGCGCCACUUACGACGCGGGAGGUCGCGGGUUCAAACCCGGAUCAGAG----------------------- --------------------..(((((.....)))))((((((......))))))..((((.(....)))))(((((....)))))........----------------------- ( -29.40, z-score = -2.15, R) >consensus _______CAGAUGCUCG_GCAUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG___________________ ........................((((((((......((((.....(((((((.......))))))).))))(((.......))).))))))))...................... (-30.02 = -29.66 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:40 2011