
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,389,623 – 14,389,718 |
| Length | 95 |
| Max. P | 0.959134 |

| Location | 14,389,623 – 14,389,718 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Shannon entropy | 0.41880 |
| G+C content | 0.45042 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14389623 95 + 27905053 CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUAA-AUUUAUGGCCC-GUCGUAAGCAA-CCAACAACUCUCAAAAUACAC- ..((((((....(....)....))))...(((.(((((((....))))))).-..((((((...-.))))))))).-))...................- ( -22.70, z-score = -1.57, R) >droSim1.chr3R 20426418 95 - 27517382 CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUAA-AUUUAUGGCCC-GUCGUAAGCAA-CCAACAACUCCCAAAAUACAC- ..((((((....(....)....))))...(((.(((((((....))))))).-..((((((...-.))))))))).-))...................- ( -22.70, z-score = -1.55, R) >droSec1.super_5 360340 95 - 5866729 CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUAA-AUUUAUGGCCC-GUCGUAAGCAA-CCGACAACUAAAGUAAUACAA- ..(((((((((.(....)....((......)).(((((((....))))))).-.))))))))).-((((.......-.))))................- ( -24.90, z-score = -2.08, R) >droYak2.chr3R 3044655 94 + 28832112 CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUAA-AUUUAUGACCC-GUCGUAAGCAA-CCAACAACUCCCA-AACACAC- ..((((((....(....)....))))...(((.(((((((....))))))).-..((((((...-.))))))))).-))...........-.......- ( -23.60, z-score = -2.26, R) >droEre2.scaffold_4770 10501772 94 - 17746568 CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUAA-AUUUAUGACCC-GUCGUAAGCAA-CCAACAGCCCCCA-AAUACUC- ..((((((....(....)....))))...(((.(((((((....))))))).-..((((((...-.))))))))).-))...........-.......- ( -23.60, z-score = -2.27, R) >droAna3.scaffold_13340 13975573 94 - 23697760 CAGGUCGUGAAAGGUAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUGA-AUUUAUGACCC-GCUGUGAGCCA-CCAACAAGACACA-GAUACAC- ..(((((((((......((((......))))..(((((((....))))))).-.))))))))).-.(((((.....-.........))))-)......- ( -24.74, z-score = -1.63, R) >dp4.chr2 18999947 96 - 30794189 CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCCAA-AUUUAUGACCCCGCCGUAAGCAAACCAACAACAUACG-GAUACGA- ..(((((((((.(....)....((......))...(((((....)))))...-.)))))))))...(((((.(...........).))))-)......- ( -22.00, z-score = -1.13, R) >droWil1.scaffold_181130 6044765 99 - 16660200 AAGGCAGUGAAAGGAAAUGUCAACCUGAGAAGGUUGGAUAAUUUAUGACCAACAGGCAUAGCCACCUUAAACGAAAACCAACGUCACGUGCAAAUUUUU ...(((((((((((.....(((((((....))))))).....(((((.((....)))))))...))))...((........)))))).)))........ ( -23.40, z-score = -1.72, R) >consensus CAGGUCGUGAAAGGAAACGUCAGCGAAGAUGCAUAGCCAAGUUUUUGGCUAA_AUUUAUGACCC_GUCGUAAGCAA_CCAACAACUCACA_AAUACAC_ ..(((((((((......((((......))))..(((((((....)))))))...))))))))).................................... (-16.85 = -17.32 + 0.47)

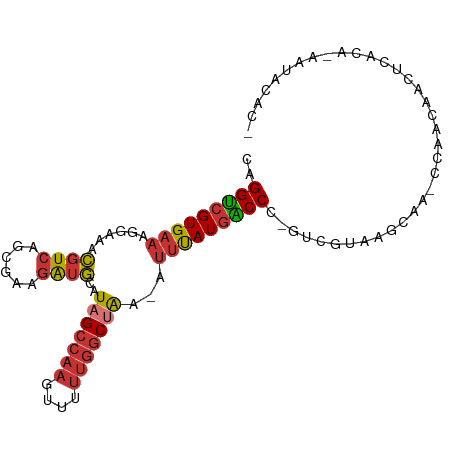
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:33 2011