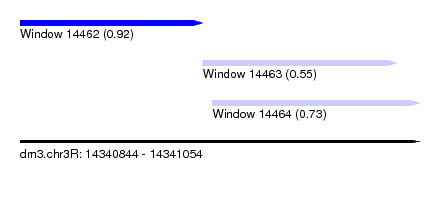
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,340,844 – 14,341,054 |
| Length | 210 |
| Max. P | 0.918753 |

| Location | 14,340,844 – 14,340,940 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.25 |
| Shannon entropy | 0.49878 |
| G+C content | 0.65842 |
| Mean single sequence MFE | -49.21 |
| Consensus MFE | -26.99 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918753 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 14340844 96 + 27905053 GAUGUUCCGCUUGUGCGGCGUCUGCAAGUCCAAGGCCGCGUAAUGCGAGUGGGCGUGGCCGUGCCCGUGGGCAUGGGCGUGGCUGUGCAGGUGCCC------------------------ ........((....))(((..(((((.......((((((((.((((......))))..(((((((....))))))))))))))).)))))..))).------------------------ ( -47.40, z-score = -1.15, R) >droSim1.chr3R 20378466 96 - 27517382 GAUGUUCCGCUUGUGCGGCGUCUGCAAGUCCAAGGCCGCGUAAUGCGAGUGGGCGUGGCCAUGCCCGUGGGCAUGGGCGUGGCUGUGCAGGUGCCC------------------------ ........((....))(((..((((..(((((....(((.....)))..)))))(..(((((((((((....)))))))))))..)))))..))).------------------------ ( -50.30, z-score = -2.05, R) >droSec1.super_5 311537 96 - 5866729 GAUGUUCCGCUUGUGCGGCGUCUGCAAGUCCAAGGCCGCAUAAUGCGAGUGGGCGUGGCCGUGCCCGUGGGCAUGGGCGUGGCUGUGCAGGUGCCC------------------------ ........((....))(((..((((..(((((....(((.....)))..)))))(..(((((((((((....)))))))))))..)))))..))).------------------------ ( -47.60, z-score = -1.24, R) >droYak2.chr3R 2985183 96 + 28832112 GAUGUUCCGCUUGUGCGGCGUCUGCAAGUCCAAGGCCGCGUAGUGCGAGUGGGCGUGGCCGUGCCCGUGGGCGUGGGCGUGGCUGUGCAUGUGCCC------------------------ ......(((((((..(.((((..((.........)).)))).)..)))))))(((..((((((((((......))))))))))..)))........------------------------ ( -44.20, z-score = -0.12, R) >droEre2.scaffold_4770 10454168 96 - 17746568 GAUGUUCCGCUUGUGCGGCGUCUGCAAGUCCAAGGCCGCGUAGUGCGAGUGGGCGUGGCCGUGCCCGUGGGCAUGGGCGUGGCUGUGCAGGUGCCC------------------------ ........((....))(((..((((..(((((....(((.....)))..)))))(..(((((((((((....)))))))))))..)))))..))).------------------------ ( -46.90, z-score = -0.86, R) >droAna3.scaffold_13340 13933478 90 - 23697760 AAUAUUCCGCUUGUGCGGUGUCUGCAAGUCCAACGCUGCAUAAUGCGAGUGGGCGUGUCCAUGCCCGUGGGC------GUGUCCAUGCAUAUGCCC------------------------ ..(((((.(((((((((((((.((......))))))))))))).))))))).(((((..((((((....)))------)))..)))))........------------------------ ( -39.10, z-score = -2.79, R) >dp4.chr2 18942957 120 - 30794189 GAUGUUGCGCUUGUGCGGCGUCUGCAAGUCGAGGGCUGCAUAGCCGUGUCCGCUGAGGGCAUGGCCAUGGCCGUGUCCGUGGCCGUGACUAUGCCCAUGGGCGUGGGUAUGCGCGUGUCC (((((((((....))))))))).((.(((.....)))(((((.(((((((((....((((((((.((((((((......)))))))).)))))))).))))))))).)))))))...... ( -62.50, z-score = -2.43, R) >droPer1.super_0 8657961 120 + 11822988 GAUGUUGCGCUUGUGCGGCGUCUGCAAGUCGAGGGCUGCAUAGCCGUGUCCGCUGAGGGCAUGGCCAUGGCCGUGUCCGUGGCCGUGACUAUGCCCAUGGGCGUGUGUAUGCGCGUGUCC (((((((((....))))))))).((((((((..(((..(.((((.......)))).(((((((((....))))))))))..))).))))).)))....(((((((((....))))))))) ( -59.40, z-score = -1.94, R) >droWil1.scaffold_181130 5986407 93 - 16660200 GAUGUUCCGCUUAUGCGGUGUCUGCAAAUCCAGUGCGGCAUACGCCGGAUGGGCAUGACCAUGGCCAUGUGCGUGGGCAUGGCCAUGGACUCC--------------------------- ........((((((.(((((((((((.......)))))...)))))).))))))....((((((((((((......)))))))))))).....--------------------------- ( -45.50, z-score = -2.63, R) >consensus GAUGUUCCGCUUGUGCGGCGUCUGCAAGUCCAAGGCCGCAUAAUGCGAGUGGGCGUGGCCAUGCCCGUGGGCAUGGGCGUGGCUGUGCAGGUGCCC________________________ ((((..((((....)))))))).(((....((.((((((......(......)...(.(((((((....))))))).))))))).))....))).......................... (-26.99 = -27.30 + 0.31)
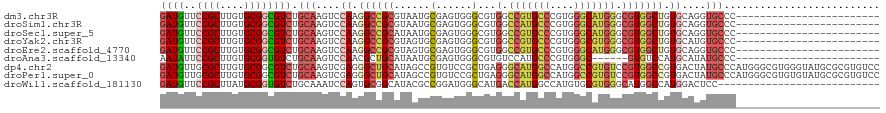
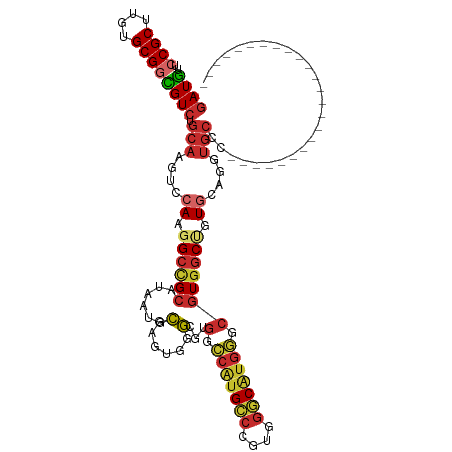
| Location | 14,340,940 – 14,341,042 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.30 |
| Shannon entropy | 0.51195 |
| G+C content | 0.62643 |
| Mean single sequence MFE | -43.71 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14340940 102 + 27905053 ------------------AUGACCGUGAGGGUGGCCCACGCCCAGCGGCGACUCGCGCGGCCGCAGAGUUGUGGGCGGUCCACGGAAAAGGGCGUAUGGAUUGCCAAAAUAAUCCUGUGA ------------------....(((((.((((.((((.......).))).)))).))))).(((((..((((.(((((((((((........)))..))))))))...))))..))))). ( -44.20, z-score = -1.24, R) >droSim1.chr3R 20378562 102 - 27517382 ------------------AUGGCCGUGAGGGUGGCCCACGCCCAGCGGCGACUCGCGCGGCCGCAGUGUUGUGGGCGGUCCGCGGAAAAGGGCGUAUGGAUUGCCAAAAUAAUCCUGUGA ------------------..(((((((.((((.((((.......).))).)))).)))))))((((..((((.(((((((((((........)))..))))))))...))))..)))).. ( -49.60, z-score = -1.72, R) >droSec1.super_5 311633 102 - 5866729 ------------------AUGGCCGUGCGGGUGGCCCACUCCCAGCGGCGACUCGCGCGGCCGCAGAGUUGUGGGCGGUCCGCGGAAUAGGGCGUAUGGAUUGCCAAAAUAAUCCUGUGA ------------------..((((((((((((.((((.......).))).))))))))))))((((..((((.(((((((((((........)))..))))))))...))))..)))).. ( -55.40, z-score = -3.31, R) >droYak2.chr3R 2985279 102 + 28832112 ------------------AUGGCCGUGGGGGUGGCCCACGCCGAGCGGCGACUCGCGCGGUCGCAGAGUUGUGGGCGGUCCGCGGAAAAGGGCGUACGGAUUGCCGAAAUAAUCCUGUGA ------------------..(((((((.((((.((((.......).))).)))).)))))))((((..((((.(((((((((((........))..)))))))))...))))..)))).. ( -47.90, z-score = -1.12, R) >droEre2.scaffold_4770 10454264 102 - 17746568 ------------------AUGGCCGUGGGGGUGGCCCACGCCGAGCGGCGACUCGCGCGGUCGCAGAGUUGUGGGCGGUCCGCGGAAGAGGGCGUACGGAUUGCCGAAAUAAUCCUGUGA ------------------..(((((((.((((.((((.......).))).)))).)))))))((((..((((.(((((((((((........))..)))))))))...))))..)))).. ( -47.90, z-score = -1.06, R) >droAna3.scaffold_13340 13933568 102 - 23697760 ------------------AUGGCCGUGGGCAUGACCGACGCCCAAUGGUGACUCCCGCGGCCUCAAAGUUGUGGGCGGACCGCGAAAAAGGGCGUAGGGAUUGCCAAAAUAAUCCUGUGA ------------------.(((.(((.((.....)).))).))).(((..(.(((((((.(((.....((((((.....))))))...))).))).)))))..))).............. ( -39.30, z-score = -0.91, R) >dp4.chr2 18943077 117 - 30794189 GUGGAUAUGGCUGUGGGCGUGGGCGUGGGCAUGGCCGACGCCUAGCGGGGACUCGCGCGGCCGCAGAGUGGUGGGUGGGCCGCGGAACAGGGCGUACGGAUUG---AAGUAAUCCUGUGA ..........((((((.(((((((((.(((...))).)))))).((((....))))))).)))))).(((((......)))))...((((((..(((......---..))).)))))).. ( -51.30, z-score = -1.20, R) >droPer1.super_0 8658081 117 + 11822988 GUGGAUAUGGCUGUGGGCGUGGGCGUGGGCAUGGCCGACGCCUAGCGGGGACUCGCGCGGCCGCAGAGUGGUGGGUGGGCCGCGGAACAGGGCGUACGGAUUG---AAGUAAUCCUGUGA ..........((((((.(((((((((.(((...))).)))))).((((....))))))).)))))).(((((......)))))...((((((..(((......---..))).)))))).. ( -51.30, z-score = -1.20, R) >droWil1.scaffold_181130 5986500 90 - 16660200 ------------------------------ACCACCCACACCCAAUGGCGACUCACGCGGUCUCAGUGUGGUGGGUGGACCGCGAAAGAGUGCAUAGGGAUUGCCAAAGUAAUCCUGUGA ------------------------------.((((((((.(.((.(((.((((.....))))))).)).)))))))))..(((......)))((((((.(((((....))))))))))). ( -34.70, z-score = -1.63, R) >droVir3.scaffold_12855 5639916 87 - 10161210 ---------------------------------AGCCACGCCCAGCGGCGAUGUGCGUGGCCGCAUUGUAAUGGGCGGGCCGCGAAAUAGAGCAUACGGAUUGCCAAAAUAAUCUUGUGA ---------------------------------.(((.((((((...((((((((......))))))))..))))))))).((........)).(((((((((......))))).)))). ( -33.40, z-score = -1.06, R) >droMoj3.scaffold_6540 25268069 87 - 34148556 ---------------------------------AGCCACGCCCAGCGGCGAACUUCGCGGCCGCAUGGUAAGGGGCGGACCGCGGAAUAGCGCAUACGGAUUGCCAAAAUAAUCCUGUGA ---------------------------------.((..((((....))))...(((((((((((..........)))).)))))))...)).((((.((((((......)))))))))). ( -33.50, z-score = -0.63, R) >droGri2.scaffold_14906 7110943 87 - 14172833 ---------------------------------AGCCACGCCCAGCGGCGAUGUGCGUGGCCGCAUUGUGAUGGGCGGGCCGCGAAAUAGUGCAUAAGGAUUGCCAAAAUAAUCCUGUGA ---------------------------------.(((.(((((((((((.((....)).))))).......)))))))))(((......)))((((.((((((......)))))))))). ( -36.01, z-score = -1.62, R) >consensus __________________AUGGCCGUGGGGGUGGCCCACGCCCAGCGGCGACUCGCGCGGCCGCAGAGUUGUGGGCGGUCCGCGGAAAAGGGCGUACGGAUUGCCAAAAUAAUCCUGUGA ..................................((..(((...)))........(((((((((..........)))).))))).....)).((((.((((((......)))))))))). (-20.50 = -20.02 + -0.48)
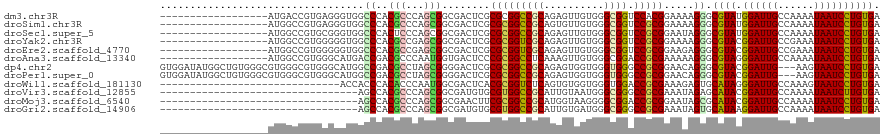
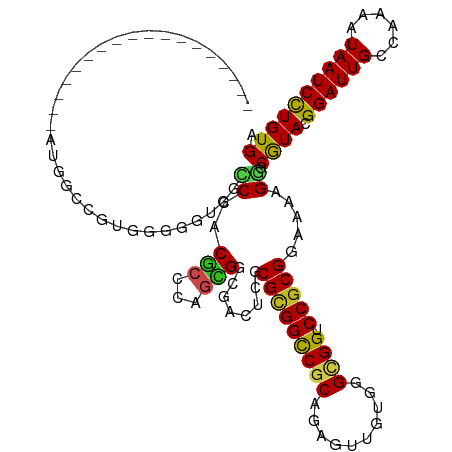
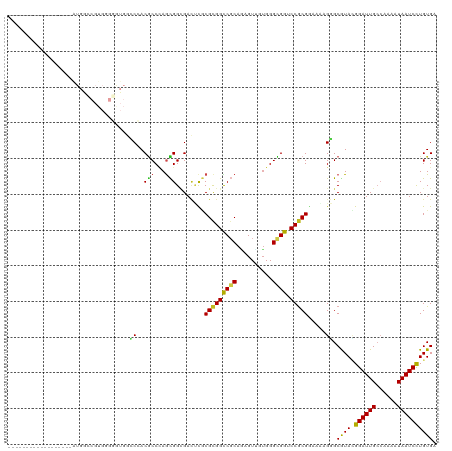
| Location | 14,340,945 – 14,341,054 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Shannon entropy | 0.44763 |
| G+C content | 0.62488 |
| Mean single sequence MFE | -47.13 |
| Consensus MFE | -25.21 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14340945 109 + 27905053 -----------CGUGAGGGUGGCCCACGCCCAGCGGCGACUCGCGCGGCCGCAGAGUUGUGGGCGGUCCACGGAAAAGGGCGUAUGGAUUGCCAAAAUAAUCCUGUGACGUCGCCAUCAU -----------.(((((((((.....)))))...((((((((((((....))((..((((.(((((((((((........)))..))))))))...))))..)))))).)))))).)))) ( -48.20, z-score = -1.50, R) >droSim1.chr3R 20378567 109 - 27517382 -----------CGUGAGGGUGGCCCACGCCCAGCGGCGACUCGCGCGGCCGCAGUGUUGUGGGCGGUCCGCGGAAAAGGGCGUAUGGAUUGCCAAAAUAAUCCUGUGACGUCGCCAUCAU -----------.(((((((((.....)))))...((((((((((((((((((..........)))).))))(((....((((.......)))).......))).)))).)))))).)))) ( -49.80, z-score = -1.37, R) >droSec1.super_5 311638 109 - 5866729 -----------CGUGCGGGUGGCCCACUCCCAGCGGCGACUCGCGCGGCCGCAGAGUUGUGGGCGGUCCGCGGAAUAGGGCGUAUGGAUUGCCAAAAUAAUCCUGUGACGUCGCCAUCAU -----------.((((((((.((((.......).))).))))))))((((((((..((((.(((((((((((........)))..))))))))...))))..))))).....)))..... ( -48.90, z-score = -1.14, R) >droYak2.chr3R 2985284 109 + 28832112 -----------CGUGGGGGUGGCCCACGCCGAGCGGCGACUCGCGCGGUCGCAGAGUUGUGGGCGGUCCGCGGAAAAGGGCGUACGGAUUGCCGAAAUAAUCCUGUGACGUCGCCAUCAU -----------.(((..((((((...(((((((......)))).)))(((((((..((((.(((((((((((........))..)))))))))...))))..)))))))))))))..))) ( -50.80, z-score = -1.29, R) >droEre2.scaffold_4770 10454269 109 - 17746568 -----------CGUGGGGGUGGCCCACGCCGAGCGGCGACUCGCGCGGUCGCAGAGUUGUGGGCGGUCCGCGGAAGAGGGCGUACGGAUUGCCGAAAUAAUCCUGUGACGUCGCCAUCAU -----------.(((..((((((...(((((((......)))).)))(((((((..((((.(((((((((((........))..)))))))))...))))..)))))))))))))..))) ( -50.80, z-score = -1.10, R) >droAna3.scaffold_13340 13933573 109 - 23697760 -----------CGUGGGCAUGACCGACGCCCAAUGGUGACUCCCGCGGCCUCAAAGUUGUGGGCGGACCGCGAAAAAGGGCGUAGGGAUUGCCAAAAUAAUCCUGUGACGUCGCCAUCAU -----------..(((((.........)))))((((((((((.((((.(((.....((((((.....))))))...))).))).(((((((......)))))))).)).))))))))... ( -41.90, z-score = -1.19, R) >dp4.chr2 18943089 117 - 30794189 GUGGGCGUGGGCGUGGGCAUGGCCGACGCCUAGCGGGGACUCGCGCGGCCGCAGAGUGGUGGGUGGGCCGCGGAACAGGGCGUACGGAUUG---AAGUAAUCCUGUGACGUGGCCAUCAU ((((.(((((((((.(((...))).)))))).((((....))))))).))))......((((...(((((((..((((((..(((......---..))).))))))..))))))).)))) ( -61.40, z-score = -3.19, R) >droPer1.super_0 8658093 117 + 11822988 GUGGGCGUGGGCGUGGGCAUGGCCGACGCCUAGCGGGGACUCGCGCGGCCGCAGAGUGGUGGGUGGGCCGCGGAACAGGGCGUACGGAUUG---AAGUAAUCCUGUGACGUGGCCAUCAU ((((.(((((((((.(((...))).)))))).((((....))))))).))))......((((...(((((((..((((((..(((......---..))).))))))..))))))).)))) ( -61.40, z-score = -3.19, R) >droWil1.scaffold_181130 5986505 97 - 16660200 -----------------------CCACACCCAAUGGCGACUCACGCGGUCUCAGUGUGGUGGGUGGACCGCGAAAGAGUGCAUAGGGAUUGCCAAAGUAAUCCUGUGAGGUGGCCAUCAU -----------------------.........(((((.(((((((((.(((...((((((......))))))..))).)))...((((((((....)))))))))))).)).)))))... ( -39.50, z-score = -2.18, R) >droVir3.scaffold_12855 5639919 96 - 10161210 ------------------------CACGCCCAGCGGCGAUGUGCGUGGCCGCAUUGUAAUGGGCGGGCCGCGAAAUAGAGCAUACGGAUUGCCAAAAUAAUCUUGUGAGGUGGCCAUCAU ------------------------..((((((...((((((((......))))))))..))))))((((((.........((((.((((((......))))))))))..))))))..... ( -37.00, z-score = -1.16, R) >droMoj3.scaffold_6540 25268072 96 - 34148556 ------------------------CACGCCCAGCGGCGAACUUCGCGGCCGCAUGGUAAGGGGCGGACCGCGGAAUAGCGCAUACGGAUUGCCAAAAUAAUCCUGUGAGGUGGCCAUCAU ------------------------..((((..((((((...(((((((((((..........)))).)))))))....)))....((((((......)))))))))..))))........ ( -35.80, z-score = -0.50, R) >droGri2.scaffold_14906 7110946 96 - 14172833 ------------------------CACGCCCAGCGGCGAUGUGCGUGGCCGCAUUGUGAUGGGCGGGCCGCGAAAUAGUGCAUAAGGAUUGCCAAAAUAAUCCUGUGAAGUGGCCAUCAU ------------------------..(((((((((((.((....)).))))).......))))))((((((.........((((.((((((......))))))))))..))))))..... ( -40.11, z-score = -2.05, R) >consensus ___________CGUGGGGGUGGCCCACGCCCAGCGGCGACUCGCGCGGCCGCAGAGUUGUGGGCGGUCCGCGGAAAAGGGCGUACGGAUUGCCAAAAUAAUCCUGUGACGUCGCCAUCAU ..................................(((.((...(((((((((..........)))).)))))........((((.((((((......))))))))))..)).)))..... (-25.21 = -24.80 + -0.41)
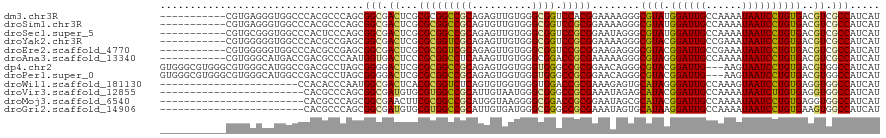
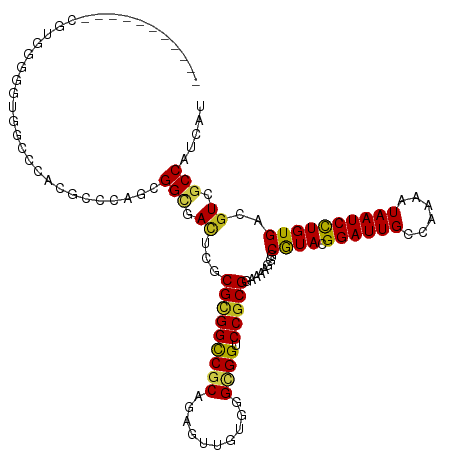
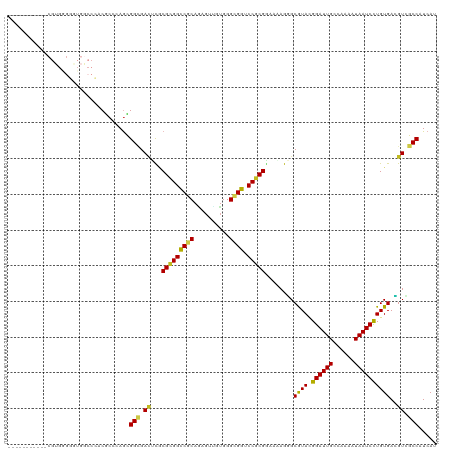
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:28 2011