| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,315,165 – 14,315,228 |
| Length | 63 |
| Max. P | 0.826335 |

| Location | 14,315,165 – 14,315,228 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.67966 |
| G+C content | 0.52566 |
| Mean single sequence MFE | -15.67 |
| Consensus MFE | -6.85 |
| Energy contribution | -7.11 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
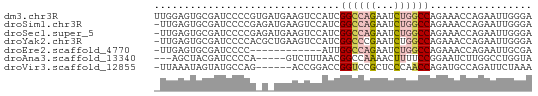
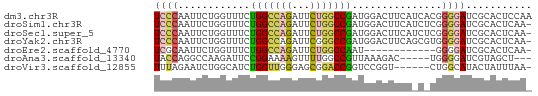
>dm3.chr3R 14315165 63 + 27905053 UUGGAGUGCGAUCCCCGUGAUGAAGUCCAUCGGCCAGAAUCUGGCCAGAAACCAGAAUUGGGA .............((((.((((.....))))((((((...))))))............)))). ( -18.80, z-score = -0.93, R) >droSim1.chr3R 20351479 62 - 27517382 -UUGAGUGCGAUCCCCGAGAUGAAGUCCAUCGGCCAGAAUCUGGCCAGAAACCAGAAUUGGGA -............(((((((((.....))))((((((...))))))...........))))). ( -20.60, z-score = -2.27, R) >droSec1.super_5 286341 62 - 5866729 -UUGAGUGCGAUCCCCGAGAUGAAGUCCAUCGGCCAGAAUCUGGCCAGAAACCAGAAUUGGGA -............(((((((((.....))))((((((...))))))...........))))). ( -20.60, z-score = -2.27, R) >droYak2.chr3R 2955928 62 + 28832112 -UUGAGUGCGAUCCCCACGCUGAAGUCCAUCGGCCCGAAUCUGGCCAGAAACCAGAAUUGGGA -....(((.(....))))(((((......)))))((((.(((((.......))))).)))).. ( -17.90, z-score = -0.86, R) >droEre2.scaffold_4770 10423673 50 - 17746568 -UUGAGUGCGAUCCCC------------AUUGGCCAGAAUCUGGCCAGAAACCAGAAUUGCGA -.....(((((((...------------.((((((((...))))))))......).)))))). ( -14.70, z-score = -2.29, R) >droAna3.scaffold_13340 13906903 55 - 23697760 ---AGCUACGAUCCCCA-----GUCUUUAACGGCCAAAACUUUUCCGGAAUCUUGGCCUGGUA ---.((((.(((.....-----)))......((((((...............)))))))))). ( -9.56, z-score = 0.57, R) >droVir3.scaffold_12855 5607109 56 - 10161210 -UUAAAUAGUAUGCCAG------ACCGGACCGGUCCGCUCCCAACCAGAUGCCAGAUUCUAAA -...........((..(------((((...))))).))........(((........)))... ( -7.50, z-score = 0.23, R) >consensus _UUGAGUGCGAUCCCCG_G_UGAAGUCCAUCGGCCAGAAUCUGGCCAGAAACCAGAAUUGGGA ...............................((((((...))))))................. ( -6.85 = -7.11 + 0.27)
| Location | 14,315,165 – 14,315,228 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.67966 |
| G+C content | 0.52566 |
| Mean single sequence MFE | -17.80 |
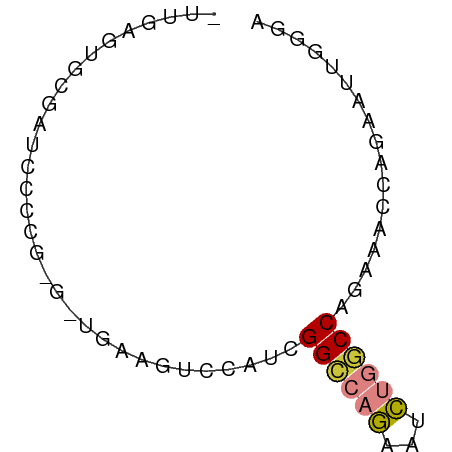
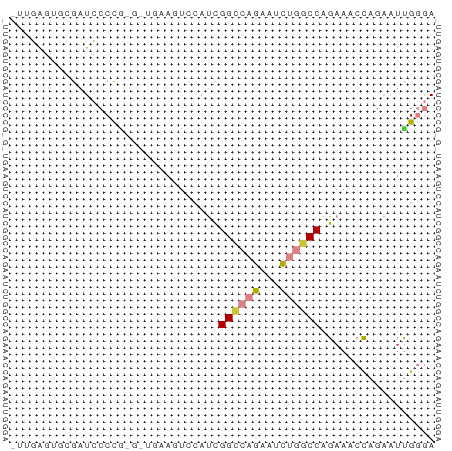
| Consensus MFE | -7.94 |
| Energy contribution | -8.19 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14315165 63 - 27905053 UCCCAAUUCUGGUUUCUGGCCAGAUUCUGGCCGAUGGACUUCAUCACGGGGAUCGCACUCCAA ((((.............((((((...))))))((((.....))))...))))........... ( -19.60, z-score = -1.07, R) >droSim1.chr3R 20351479 62 + 27517382 UCCCAAUUCUGGUUUCUGGCCAGAUUCUGGCCGAUGGACUUCAUCUCGGGGAUCGCACUCAA- ((((.............((((((...))))))((((.....))))...))))..........- ( -19.10, z-score = -1.04, R) >droSec1.super_5 286341 62 + 5866729 UCCCAAUUCUGGUUUCUGGCCAGAUUCUGGCCGAUGGACUUCAUCUCGGGGAUCGCACUCAA- ((((.............((((((...))))))((((.....))))...))))..........- ( -19.10, z-score = -1.04, R) >droYak2.chr3R 2955928 62 - 28832112 UCCCAAUUCUGGUUUCUGGCCAGAUUCGGGCCGAUGGACUUCAGCGUGGGGAUCGCACUCAA- (((((...((((..(((((((.......))))...)))..))))..)))))...........- ( -19.20, z-score = -0.19, R) >droEre2.scaffold_4770 10423673 50 + 17746568 UCGCAAUUCUGGUUUCUGGCCAGAUUCUGGCCAAU------------GGGGAUCGCACUCAA- ..((.(((((.((...(((((((...)))))))))------------.))))).))......- ( -18.50, z-score = -2.60, R) >droAna3.scaffold_13340 13906903 55 + 23697760 UACCAGGCCAAGAUUCCGGAAAAGUUUUGGCCGUUAAAGAC-----UGGGGAUCGUAGCU--- ..(((((((((((((.......))))))))))(((...)))-----)))...........--- ( -14.90, z-score = -0.78, R) >droVir3.scaffold_12855 5607109 56 + 10161210 UUUAGAAUCUGGCAUCUGGUUGGGAGCGGACCGGUCCGGU------CUGGCAUACUAUUUAA- ..(((..((..((.....))..))..(((((((...))))------))).....))).....- ( -14.20, z-score = -0.18, R) >consensus UCCCAAUUCUGGUUUCUGGCCAGAUUCUGGCCGAUGGACUUCA_C_CGGGGAUCGCACUCAA_ ((((............(((((((...)))))))...............))))........... ( -7.94 = -8.19 + 0.25)
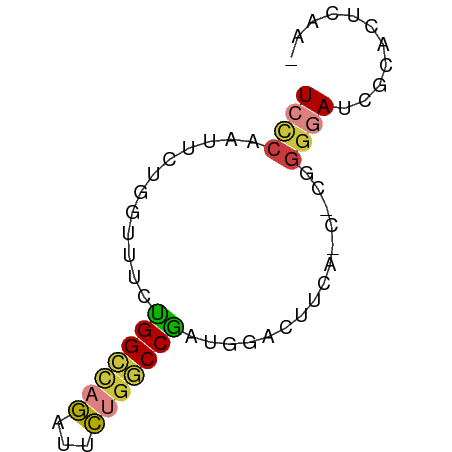
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:23 2011