| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,286,693 – 14,286,800 |
| Length | 107 |
| Max. P | 0.625333 |

| Location | 14,286,693 – 14,286,800 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
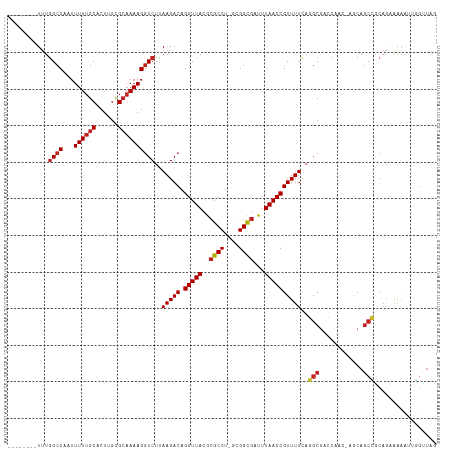
| Mean pairwise identity | 84.06 |
| Shannon entropy | 0.33754 |
| G+C content | 0.48701 |
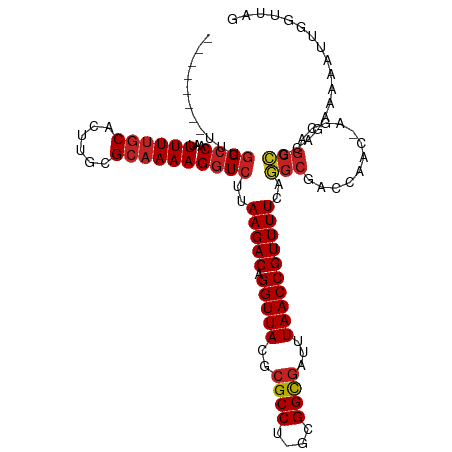
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14286693 107 + 27905053 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU-GUGGCGAUUUAACCGUUUUCAGGCGACCAAC-GGCAGCCGCAGAAAAAUUGGUUAG --------..((((((((((((......((((.....(((....))).(....)))))((-(((((.......(((((.((....))..)))-))..))))))))))))))))))). ( -35.80, z-score = -1.95, R) >droEre2.scaffold_4770 10394374 107 - 17746568 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCAGGCGACCAAC-AGCAGCCGCAGAAAAAUUGGUUAG --------..((((((((((((......((((.....(((....))).(....)))))((-(((((((...........))..((.......-.)).))))))))))))))))))). ( -36.00, z-score = -2.25, R) >droYak2.chr3R 2919916 107 + 28832112 --------UUUGGACAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCUGGCGACCAAC-GGCAGCCGCAGGAAAAUUGGAUAG --------..((..(((((((.......(((.((...(((....)))...)).))).(((-(((((.......(((((.((....))..)))-))..)))))))))))))))..)). ( -33.10, z-score = -1.29, R) >droSec1.super_5 258053 107 - 5866729 --------UUUAGCCAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU-GCGGCAAUUUAACCGUUUUCAGGCGACCAAC-GGCAGCCGCAGAAAAAUUGGUUAG --------..((((((((((((......((((.....(((....))).(....)))))((-(((((.......(((((.((....))..)))-))..))))))))))))))))))). ( -37.00, z-score = -2.88, R) >droSim1.chr3R 20321160 107 - 27517382 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCAGGCGACCAAC-GGCAGCCGCAGAAAAAUUGGUUAG --------..((((((((((((......((((.....(((....))).(....)))))((-(((((.......(((((.((....))..)))-))..))))))))))))))))))). ( -37.70, z-score = -2.38, R) >dp4.chr2 18886728 107 - 30794189 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUCAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCAUGCGAGCGGC-CACAGCAGCAACAAAGAUUGCUAG --------..(((((...((((((......))))))(.((.((.((..(((((..((((.-..))))...)))))....)).)).)).))))-))....(((((......))))).. ( -33.50, z-score = -1.04, R) >droPer1.super_0 8600496 107 + 11822988 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUCAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCAUGCGAGCGGC-CACAGCAGCAACAAAGAUUGCUAG --------..(((((...((((((......))))))(.((.((.((..(((((..((((.-..))))...)))))....)).)).)).))))-))....(((((......))))).. ( -33.50, z-score = -1.04, R) >droWil1.scaffold_181130 5918869 106 - 16660200 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUCAAGACAGGUUACGCGCCUAGAGGCGACUUAACCGUUUUCAUGCGAACAACAAACA--UAAAGGGAAGCUGCUUG- --------...((((...((((((......)))))))))).((((.(((((((.(((((....)))).).)))))((((((................--.....))))))))))))- ( -27.50, z-score = -0.66, R) >droAna3.scaffold_13340 13878752 106 - 23697760 --------UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUCAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCAGGCGGCCAGA-AGCAGCCACAAAAAAAU-GGUUAG --------(((((((..(((((((......)))))))((((.(((((.(((((..((((.-..))))...)))))))))).)))))))))))-...(((((........)-)))).. ( -39.60, z-score = -3.25, R) >droMoj3.scaffold_6540 25187804 115 - 34148556 CUGCCAGUUUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU-GCGGCGAUUUAACCGUUUUCAUGCAACAAAU-ACCAGCAACAACAACAACUGUUCG ....(((((..((((...((((((......))))))))))..(((((.(((((..((((.-..))))...))))))))))..(((.......-....)))........))))).... ( -25.80, z-score = 0.22, R) >droGri2.scaffold_14906 7052159 97 - 14172833 --------UCUGGCCAAUUUUUGCACUUGUGGUAAAGGUCUUAAGACAGGUUACGCGCCU-GCGGUGAUCUAACCGUUUUUAUGCAAGCCGCUAACAGCAGCCGCA----------- --------..(((((...((((((.(....)))))))(((....))).))))).(((.((-(((((......)))(((.....((.....)).))).)))).))).----------- ( -26.30, z-score = 0.50, R) >consensus ________UUUGGCCAAUUUUUGCACUUGCGCAAAAGGUCUUAAGACAGGUUACGCGCCU_GCGGCGAUUUAACCGUUUUCAGGCGACCAAC_AGCAGCCGCAGAAAAAUUGGUUAG ...........((((...((((((......))))))))))..(((((.(((((..((((....))))...))))))))))..(((............)))................. (-23.14 = -23.39 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:17 2011