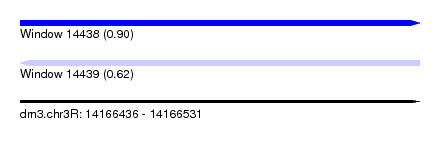
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,166,436 – 14,166,531 |
| Length | 95 |
| Max. P | 0.900770 |

| Location | 14,166,436 – 14,166,531 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Shannon entropy | 0.36912 |
| G+C content | 0.40172 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.07 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
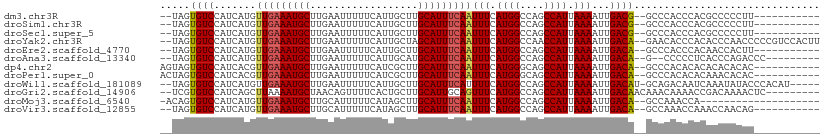
>dm3.chr3R 14166436 95 + 27905053 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACG--GCCCACCCACGCCCCCUU----------- --..(((.((((...((((((((((..((((.....))).)..))))))))))...))))...(((.(((....))).)--)).....)))........----------- ( -21.10, z-score = -1.76, R) >droSim1.chr3R 20205616 95 - 27517382 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACG--GCCCACCCACGCCCCCUU----------- --..(((.((((...((((((((((..((((.....))).)..))))))))))...))))...(((.(((....))).)--)).....)))........----------- ( -21.10, z-score = -1.76, R) >droSec1.super_5 134680 95 - 5866729 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACG--GCCCACCCACGCCCCCUU----------- --..(((.((((...((((((((((..((((.....))).)..))))))))))...))))...(((.(((....))).)--)).....)))........----------- ( -21.10, z-score = -1.76, R) >droYak2.chr3R 2776953 106 + 28832112 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUAGCAUUUCAAUUUCAUGGCCAACCAUUAAAAUUGACA--GAACACCCACACCCAACCCCCGUCCACUU --.((((.((((...((((((((((((((.....))).....)))))))))))...))))...............(((.--.....................))))))). ( -17.85, z-score = -1.59, R) >droEre2.scaffold_4770 10275085 95 - 17746568 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACA--GCCCACCCACAACCACUU----------- --..(((.((((...((((((((((..((((.....))).)..))))))))))...))))...((..(((....)))..--)).....)))........----------- ( -17.20, z-score = -0.81, R) >droAna3.scaffold_13340 13763429 95 - 23697760 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCAUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACA--G--CCCCCUCACCCAGACCC--------- --...((((......((((((((((.(((((.....))).)).))))))))))...((((....)))).......))))--.--.................--------- ( -17.70, z-score = -0.99, R) >dp4.chr2 18753279 97 - 30794189 AGUAGUGUCCAUCACGUUGAAAUGCUUGAAUUUUUCAUCGCUUGCAUUUCAAUUUCAUGGGCAGCCAUUAAAAUUGACA--GCCCACACACACACACAC----------- .((.((((.......((((((((((.(((........)))...))))))))))....(((((.(.((.......)).).--))))).))))))......----------- ( -23.30, z-score = -2.39, R) >droPer1.super_0 8457994 97 + 11822988 ACUAGUGUCCAUCACGUUGAAAUGCUUGAAUUUUUCAUCGCUUGCAUUUCAAUUUCAUGGGCAGCCAUUAAAAUUGACA--GCCCACACACAAACACAC----------- ....((((.......((((((((((.(((........)))...))))))))))....(((((.(.((.......)).).--))))).))))........----------- ( -22.80, z-score = -2.61, R) >droWil1.scaffold_181089 1013496 102 + 12369635 --UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAUUUUCAUGGCCAGCCAUUAAAAUUGACAU-GCAGACAAUCAAAUAUACCCACAU----- --...((((...(((((((((((((..((((.....))).)..)))))))(((((.((((....)))).))))).)))))-)..)))).................----- ( -22.80, z-score = -1.75, R) >droGri2.scaffold_14906 6931292 99 - 14172833 --UCGUGUCCAUCAGCUUAAAAUGCUAACAGUUUUCACUGCUUGCAUUGCAGUUUCAUGGCCAGCCAUUAAAAUUGACAACAAACAAAACCGACAAAACUC--------- --...((((....(((.......)))..(((((((.(((((.......)))))...((((....)))).)))))))...............))))......--------- ( -18.40, z-score = -1.12, R) >droMoj3.scaffold_6540 25052222 87 - 34148556 -ACAGUGUCCAUCAUGUUGAAAUGCUUGCAUUUUUCAUAGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACA--GCCAAACCA-------------------- -....((((......((((((((((..((..........))..))))))))))...((((....)))).......))))--.........-------------------- ( -20.40, z-score = -1.75, R) >droVir3.scaffold_12855 5431328 95 - 10161210 --UAGUGUCCAUCAUGUUGAAAUGCUUGCAUUUUUCAUAGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACA--GCCAAACCAAACCAACAG----------- --...((((......((((((((((..((..........))..))))))))))...((((....)))).......))))--..................----------- ( -20.40, z-score = -1.86, R) >consensus __UAGUGUCCAUCAUGUUGAAAUGCUUGAAUUUUUCAUUGCUUGCAUUUCAAUUUCAUGGCCAGCCAUUAAAAUUGACA__GCCCACCCACACCCACAC___________ .....((.((((...((((((((((..................))))))))))...)))).))............................................... (-14.81 = -15.07 + 0.26)
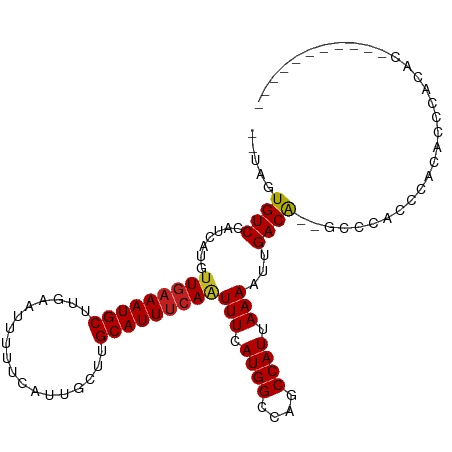
| Location | 14,166,436 – 14,166,531 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Shannon entropy | 0.36912 |
| G+C content | 0.40172 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14166436 95 - 27905053 -----------AAGGGGGCGUGGGUGGGC--CGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- -----------.......(((((((.(((--(((........)))))).)))......(((((((((..(.(((.....))))..)))))))))))))..........-- ( -26.70, z-score = -1.61, R) >droSim1.chr3R 20205616 95 + 27517382 -----------AAGGGGGCGUGGGUGGGC--CGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- -----------.......(((((((.(((--(((........)))))).)))......(((((((((..(.(((.....))))..)))))))))))))..........-- ( -26.70, z-score = -1.61, R) >droSec1.super_5 134680 95 + 5866729 -----------AAGGGGGCGUGGGUGGGC--CGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- -----------.......(((((((.(((--(((........)))))).)))......(((((((((..(.(((.....))))..)))))))))))))..........-- ( -26.70, z-score = -1.61, R) >droYak2.chr3R 2776953 106 - 28832112 AAGUGGACGGGGGUUGGGUGUGGGUGUUC--UGUCAAUUUUAAUGGUUGGCCAUGAAAUUGAAAUGCUAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- .(.(.(((....))).).)...(((((((--.(((((((.....)))))))((((...((((((((((.(.(((.....)))).))))))))))))))..))))))).-- ( -28.50, z-score = -1.65, R) >droEre2.scaffold_4770 10275085 95 + 17746568 -----------AAGUGGUUGUGGGUGGGC--UGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- -----------.((((((..(((((.(((--(((........)))))).)))......(((((((((..(.(((.....))))..)))))))))))..))...)))).-- ( -26.80, z-score = -1.87, R) >droAna3.scaffold_13340 13763429 95 + 23697760 ---------GGGUCUGGGUGAGGGGG--C--UGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAUGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- ---------(((((((.(((....((--(--(((((.......)))).))))......(((((((((.((.(((.....))))).))))))))))))..))))).)).-- ( -26.50, z-score = -1.64, R) >dp4.chr2 18753279 97 + 30794189 -----------GUGUGUGUGUGUGUGGGC--UGUCAAUUUUAAUGGCUGCCCAUGAAAUUGAAAUGCAAGCGAUGAAAAAUUCAAGCAUUUCAACGUGAUGGACACUACU -----------(((.((((.(((((((((--.((((.......)))).))))))....(((((((((....((........))..)))))))))....))).))))))). ( -29.70, z-score = -2.44, R) >droPer1.super_0 8457994 97 - 11822988 -----------GUGUGUUUGUGUGUGGGC--UGUCAAUUUUAAUGGCUGCCCAUGAAAUUGAAAUGCAAGCGAUGAAAAAUUCAAGCAUUUCAACGUGAUGGACACUAGU -----------..(((((..(..((((((--.((((.......)))).))))))....(((((((((....((........))..)))))))))....)..))))).... ( -30.50, z-score = -2.73, R) >droWil1.scaffold_181089 1013496 102 - 12369635 -----AUGUGGGUAUAUUUGAUUGUCUGC-AUGUCAAUUUUAAUGGCUGGCCAUGAAAAUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA-- -----..(((......(((.((((..(((-((.(((.(((((.(((....)))))))).))).)))))..)))).)))........((((.......))))..)))..-- ( -25.10, z-score = -1.09, R) >droGri2.scaffold_14906 6931292 99 + 14172833 ---------GAGUUUUGUCGGUUUUGUUUGUUGUCAAUUUUAAUGGCUGGCCAUGAAACUGCAAUGCAAGCAGUGAAAACUGUUAGCAUUUUAAGCUGAUGGACACGA-- ---------..((((..(((((((.....((((.((.(((((.(((....)))))))).))))))((.((((((....)))))).)).....)))))))..)).))..-- ( -29.30, z-score = -1.88, R) >droMoj3.scaffold_6540 25052222 87 + 34148556 --------------------UGGUUUGGC--UGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCUAUGAAAAAUGCAAGCAUUUCAACAUGAUGGACACUGU- --------------------....(..((--(((........)))))..)((((.(..(((((((((..((..........))..)))))))))..).)))).......- ( -22.00, z-score = -1.03, R) >droVir3.scaffold_12855 5431328 95 + 10161210 -----------CUGUUGGUUUGGUUUGGC--UGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCUAUGAAAAAUGCAAGCAUUUCAACAUGAUGGACACUA-- -----------.((((.(((((...((((--(((((.......)))).))))).....(((((((((..((..........))..))))))))))).))).))))...-- ( -25.90, z-score = -1.67, R) >consensus ___________GUGUGGGUGUGGGUGGGC__UGUCAAUUUUAAUGGCUGGCCAUGAAAUUGAAAUGCAAGCAAUGAAAAAUUCAAGCAUUUCAACAUGAUGGACACUA__ ................................((((((((((.(((....))))))))))(((((((..(.(((.....))))..))))))).........)))...... (-15.61 = -15.45 + -0.16)

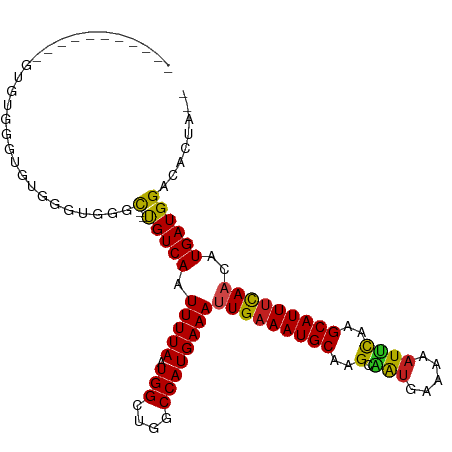
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:07 2011