| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,165,744 – 14,165,869 |
| Length | 125 |
| Max. P | 0.974102 |

| Location | 14,165,744 – 14,165,869 |
|---|---|
| Length | 125 |
| Sequences | 12 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 48.66 |
| Shannon entropy | 1.13184 |
| G+C content | 0.48726 |
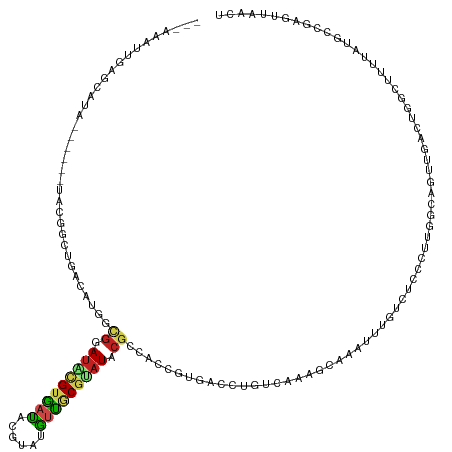
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -6.27 |
| Energy contribution | -6.34 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.92 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
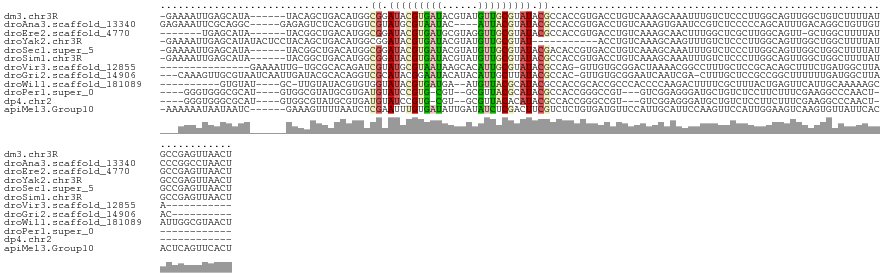
>dm3.chr3R 14165744 125 - 27905053 -GAAAAUUGAGCAUA------UACAGCUGACAUGGCGGAUACGUGAUACGUAUGUUGCGUAUACGCCACCGUGACCUGUCAAAGCAAAUUUGUCUCCCUUGGCAGUUGGCUGUCUUUUAUGCCGAGUUAACU -.....(((.(((((------.(((((..((.(((((.(((((..(((....)))..))))).)))))........((((((((((....)).))...))))))))..)))))....))))))))....... ( -41.50, z-score = -2.99, R) >droAna3.scaffold_13340 13762728 123 + 23697760 GAGAAAUUCGCAGGC-----GAGAGUCUCACGUGUCGUAUGCGUAAUAC----AUUACGUAUACGCCACCGUGACCUGUCAAAGUGAAUCCGUCUCCCCCAGCAUUUGACAGGCUGUUGUCCCGGCCUAACU ((((.((((((.(((-----....)))(((((((.(((((((((((...----.))))))))))).)).))))).........))))))...))))..............((((((......)))))).... ( -44.00, z-score = -4.19, R) >droEre2.scaffold_4770 10274463 118 + 17746568 -------UGAGCAUA------UACGGCUGACAUGGCGGAUACGUGAUGCGUAGGUUGCGUAUACGCCACCGUGACCUGUCAAAGCAACUUUGGCUCGCUUGGCAGUU-GCUGGCUUUUAUGCCGAGUUAACU -------.((((...------(((((......(((((.(((((..((......))..))))).))))))))))......(((((...)))))))))((((((((...-(....).....))))))))..... ( -40.90, z-score = -0.98, R) >droYak2.chr3R 2776287 120 - 28832112 -GAAAAUUGAGCAUAUACUCCUACAGCUGACAUGGCGGAUACGUGAUACGUAUGUUGCGUAU-----------ACCUGUCAAAGCAAGUUUGUCUCCCUUGGCAGUUGGCUGGCUUUUAUGCCGAGUUAACU -.....(((.(((((........((((..((.(((((((((((..(((....)))..)))))-----------.)).)))).........((((......))))))..)))).....))))))))....... ( -36.32, z-score = -1.72, R) >droSec1.super_5 133991 125 + 5866729 -GAAAAUUGAGCAUA------UACGGCUGACAUGGCGGAUACGUGAUACGUAUGUUGCGUAUACGACACCGUGACCUGUCAAAGCAAAUUUGUCUCCCUUGGCAGUUGGCUGGCUUUUAUGCCGAGUUAACU -.....(((.(((((------..((((..((.(((((((((((..(((....)))..)))))(((....)))...)))))).........((((......))))))..)))).....))))))))....... ( -37.50, z-score = -1.48, R) >droSim1.chr3R 20204929 125 + 27517382 -GAAAAUUGAGCAUA------UACGGCUGACAUGGCGGAUACGUGAUACGUAUGUUGCGUAUACGCCACCGUGACCUGUCAAAGCAAAUUUGUCUCCCUUGGCAGUUGGCUGGCUUUUAUGCCGAGUUAACU -..(((((..((...------.((((.(.((.(((((.(((((..(((....)))..))))).)))))..)).).))))....)).)))))(((......)))(((((((((((......))).)))))))) ( -40.50, z-score = -2.08, R) >droVir3.scaffold_12855 5429576 104 + 10161210 ---------------GAAAAUUG-UGCGCACAGAUCGUAUGCGUAAUAAGCACAUUGCGUAUACGCCAG-GUUGUGCGGACUAAAACGGCCUUUGCUCCGCACAGCUUUCUGAUGGCUUAA----------- ---------------........-.((.((((((.((((((((((((......))))))))))))..((-((((((((((.((((......)))).)))))))))))))))).))))....----------- ( -40.50, z-score = -3.53, R) >droGri2.scaffold_14906 6930284 117 + 14172833 ---CAAAGUUGCGUAAUCAAUUGAUACGCACAGGUCGCAUACGGAAUACAUACAUUGCUUAUACGCCAC-GUUGUGCGGAAUCAAUCGA-CUUUGCUCCGCCGGCUUUUUUGAUGGCUUAAC---------- ---((((((((........((((((.(((((((((.(((((.(.(((......))).).)))..)).))-.)))))))..)))))))))-)))))....((((.(......).)))).....---------- ( -28.50, z-score = -0.36, R) >droWil1.scaffold_181089 1012265 115 - 12369635 ----------GUGUAU----GC-UUGUAUACGUGUGGUAUACGUGAUGA--AUGUUACGCAUACGCCACCGCACCGCCCACCCCAAGACUUUUCGCUUUACUGAGUUCAUUGCAAAAAGCAUUGGCGUAACU ----------((((((----((-..((((((.....))))))(((((..--..))))))))))))).............((.((((..(((((.((...............)).)))))..)))).)).... ( -30.56, z-score = -0.86, R) >droPer1.super_0 8456508 105 - 11822988 ----GGGUGGGCGCAU----GUGGCGUAUGCGUGAUGUAUCCGUG-CGU--GCGUUACGCAUACGCCACCGGGCCGU---GUCGGAGGGAUGCUGUCUCCUUCUUUCGAAGGCCCAACU------------- ----((((.(((.(..----((((((((((((((((((((.....-.))--)))))))))))))))))).).))).(---(..(((((((.......)))))))..))...))))....------------- ( -55.60, z-score = -4.02, R) >dp4.chr2 18751779 105 + 30794189 ----GGGUGGGCGCAU----GUGGCGUAUGCGUGAUGUAUCCGUG-CGU--GCGUUACACAUACGCCACCGGGCCGU---GUCGGAGGGAUGCUGUCUCCUUCUUUCGAAGGCCCAACU------------- ----.(((((.((.((----((((((((((((((.......))))-)))--))))..))))).)))))))(((((..---.(((((((((.(......).))))))))).)))))....------------- ( -50.60, z-score = -2.85, R) >apiMel3.Group10 9206558 125 + 11440700 -AAAAAAUAAUAAUC------GAAAGUUUUAAUCUCGAAUUUGUGAUAUUGAUAUCUCGACUUCGUCUCUGUGAUGUUCCAUUGCAUUCCAAGUUCCAUUGGAAGUCAAGUGUUAUUCACACUCAGUUCACU -.........(((.(------....)..))).....((((((((((...(((((.((.((((((.....((..(((...)))..))...(((......))))))))).))))))).)))))...)))))... ( -24.80, z-score = -1.46, R) >consensus ___AAAUUGAGCAUA______UACGGCUGACAUGGCGGAUACGUGAUACGUAUGUUGCGUAUACGCCACCGUGACCUGUCAAAGCAAAUUUGUCUCCCUUGGCAGUUGACUGGCUUUUAUGCCGAGUUAACU ...................................((.((((((((((....)))))))))).))................................................................... ( -6.27 = -6.34 + 0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:05 2011